[English] 日本語
 Yorodumi
Yorodumi- EMDB-13902: The cryoEM density map of T20s proteasome with various ice thickn... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13902 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The cryoEM density map of T20s proteasome with various ice thickness subsets 4 and 5 combined (EMPIAR-10025 reprocessing) | |||||||||||||||
 Map data Map data | Sharpened and masked map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |   Thermoplasma acidophilum (acidophilic) Thermoplasma acidophilum (acidophilic) | |||||||||||||||
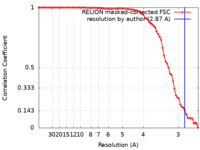
| Method | single particle reconstruction / cryo EM / Resolution: 2.87 Å | |||||||||||||||
 Authors Authors | Olek M / Zhang P / Cowtan K / Chaban Y / Webb D | |||||||||||||||
| Funding support | European Union, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2022 Journal: Structure / Year: 2022Title: IceBreaker: Software for high-resolution single-particle cryo-EM with non-uniform ice. Authors: Mateusz Olek / Kevin Cowtan / Donovan Webb / Yuriy Chaban / Peijun Zhang /  Abstract: Despite the abundance of available software tools, optimal particle selection is still a vital issue in single-particle cryoelectron microscopy (cryo-EM). Regardless of the method used, most pickers ...Despite the abundance of available software tools, optimal particle selection is still a vital issue in single-particle cryoelectron microscopy (cryo-EM). Regardless of the method used, most pickers struggle when ice thickness varies on a micrograph. IceBreaker allows users to estimate the relative ice gradient and flatten it by equalizing the local contrast. It allows the differentiation of particles from the background and improves overall particle picking performance. Furthermore, we introduce an additional parameter corresponding to local ice thickness for each particle. Particles with a defined ice thickness can be grouped and filtered based on this parameter during processing. These functionalities are especially valuable for on-the-fly processing to automatically pick as many particles as possible from each micrograph and to select optimal regions for data collection. Finally, estimated ice gradient distributions can be stored separately and used to inspect the quality of prepared samples. #1:  Journal: Elife / Year: 2015 Journal: Elife / Year: 2015Title: 2.8 A resolution reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy. Authors: Campbell MG / Veesler D / Cheng A / Potter CS / Carragher B | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13902.map.gz emd_13902.map.gz | 5.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13902-v30.xml emd-13902-v30.xml emd-13902.xml emd-13902.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13902_fsc.xml emd_13902_fsc.xml | 7.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_13902.png emd_13902.png | 166.6 KB | ||
| Masks |  emd_13902_msk_1.map emd_13902_msk_1.map | 38.4 MB |  Mask map Mask map | |
| Others |  emd_13902_additional_1.map.gz emd_13902_additional_1.map.gz emd_13902_additional_2.map.gz emd_13902_additional_2.map.gz emd_13902_half_map_1.map.gz emd_13902_half_map_1.map.gz emd_13902_half_map_2.map.gz emd_13902_half_map_2.map.gz | 36 MB 29.5 MB 29.6 MB 29.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13902 http://ftp.pdbj.org/pub/emdb/structures/EMD-13902 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13902 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13902 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13902.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13902.map.gz / Format: CCP4 / Size: 38.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened and masked map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.315 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
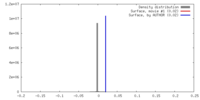
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13902_msk_1.map emd_13902_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
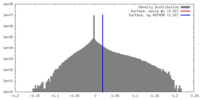
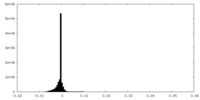
| Density Histograms |
-Additional map: Sharpened map (not masked)
| File | emd_13902_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map (not masked) | ||||||||||||
| Projections & Slices |
| ||||||||||||
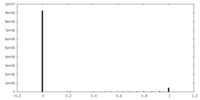
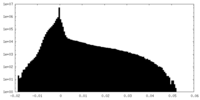
| Density Histograms |
-Additional map: Unsharpened map
| File | emd_13902_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
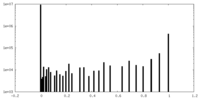
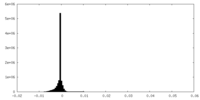
| Density Histograms |
-Half map: Half map 1
| File | emd_13902_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_13902_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
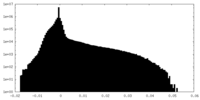
| Density Histograms |
- Sample components
Sample components
-Entire : Thermoplasma acidophilum 20S proteasome
| Entire | Name: Thermoplasma acidophilum 20S proteasome |
|---|---|
| Components |
|
-Supramolecule #1: Thermoplasma acidophilum 20S proteasome
| Supramolecule | Name: Thermoplasma acidophilum 20S proteasome / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Thermoplasma acidophilum (acidophilic) Thermoplasma acidophilum (acidophilic) |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.21 mg/mL |
|---|---|
| Buffer | pH: 7.8 / Details: 20 mM Tris, 150 mM NaCl |
| Vitrification | Cryogen name: ETHANE / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 53.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)