[English] 日本語
 Yorodumi
Yorodumi- EMDB-13867: Structure of the bacterial type VI secretion system effector RhsA. -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13867 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the bacterial type VI secretion system effector RhsA. | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | bacterial type VI secretion system / effector / Rhs repeats / TOXIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||
| Biological species |  Pseudomonas protegens Pf-5 (bacteria) Pseudomonas protegens Pf-5 (bacteria) | ||||||||||||
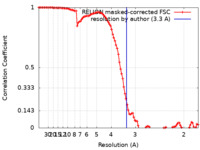
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | ||||||||||||
 Authors Authors | Guenther P / Quentin D | ||||||||||||
| Funding support | 3 items
| ||||||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2022 Journal: PLoS Pathog / Year: 2022Title: Structure of a bacterial Rhs effector exported by the type VI secretion system. Authors: Patrick Günther / Dennis Quentin / Shehryar Ahmad / Kartik Sachar / Christos Gatsogiannis / John C Whitney / Stefan Raunser /   Abstract: The type VI secretion system (T6SS) is a widespread protein export apparatus found in Gram-negative bacteria. The majority of T6SSs deliver toxic effector proteins into competitor bacteria. Yet, the ...The type VI secretion system (T6SS) is a widespread protein export apparatus found in Gram-negative bacteria. The majority of T6SSs deliver toxic effector proteins into competitor bacteria. Yet, the structure, function, and activation of many of these effectors remains poorly understood. Here, we present the structures of the T6SS effector RhsA from Pseudomonas protegens and its cognate T6SS spike protein, VgrG1, at 3.3 Å resolution. The structures reveal that the rearrangement hotspot (Rhs) repeats of RhsA assemble into a closed anticlockwise β-barrel spiral similar to that found in bacterial insecticidal Tc toxins and in metazoan teneurin proteins. We find that the C-terminal toxin domain of RhsA is autoproteolytically cleaved but remains inside the Rhs 'cocoon' where, with the exception of three ordered structural elements, most of the toxin is disordered. The N-terminal 'plug' domain is unique to T6SS Rhs proteins and resembles a champagne cork that seals the Rhs cocoon at one end while also mediating interactions with VgrG1. Interestingly, this domain is also autoproteolytically cleaved inside the cocoon but remains associated with it. We propose that mechanical force is required to remove the cleaved part of the plug, resulting in the release of the toxin domain as it is delivered into a susceptible bacterial cell by the T6SS. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13867.map.gz emd_13867.map.gz | 85.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13867-v30.xml emd-13867-v30.xml emd-13867.xml emd-13867.xml | 18.9 KB 18.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13867_fsc.xml emd_13867_fsc.xml | 10.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_13867.png emd_13867.png | 65.9 KB | ||
| Masks |  emd_13867_msk_1.map emd_13867_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13867.cif.gz emd-13867.cif.gz | 6.8 KB | ||
| Others |  emd_13867_half_map_1.map.gz emd_13867_half_map_1.map.gz emd_13867_half_map_2.map.gz emd_13867_half_map_2.map.gz | 70.1 MB 70.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13867 http://ftp.pdbj.org/pub/emdb/structures/EMD-13867 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13867 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13867 | HTTPS FTP |
-Related structure data
| Related structure data |  7q97MC  7q5pC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13867.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13867.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.91 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13867_msk_1.map emd_13867_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_13867_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_13867_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of RhsA.
| Entire | Name: Structure of RhsA. |
|---|---|
| Components |
|
-Supramolecule #1: Structure of RhsA.
| Supramolecule | Name: Structure of RhsA. / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Pseudomonas protegens Pf-5 (bacteria) Pseudomonas protegens Pf-5 (bacteria) |
| Molecular weight | Theoretical: 164.91297 KDa |
-Macromolecule #1: Rhs family protein
| Macromolecule | Name: Rhs family protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas protegens Pf-5 (bacteria) / Strain: ATCC BAA-477 / NRRL B-23932 / Pf-5 Pseudomonas protegens Pf-5 (bacteria) / Strain: ATCC BAA-477 / NRRL B-23932 / Pf-5 |
| Molecular weight | Theoretical: 159.719453 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SQDPSKTGAD KGLSNLCEGI ANALFPPSIQ ATIASGSKKV LTNNKPAARA AGIAPPTQIS GDGAEEGEAT DTPSATPEE GPGFLDMAKE FFSQMWRPTV ASPAPGSQTK ELDLVTCTKH PPMPVQYLAE GSEKVSIDGQ PAVRSGDRST C DAKVVSAG ...String: MGSSHHHHHH SQDPSKTGAD KGLSNLCEGI ANALFPPSIQ ATIASGSKKV LTNNKPAARA AGIAPPTQIS GDGAEEGEAT DTPSATPEE GPGFLDMAKE FFSQMWRPTV ASPAPGSQTK ELDLVTCTKH PPMPVQYLAE GSEKVSIDGQ PAVRSGDRST C DAKVVSAG PISPNVTIGG GSVVVREIRS GKTPGVGLAV TVLMMLKGGK GKFLSNLPCM LMAGVNSFVV SQATNALTQA VV ASTHPVH AATGAKVLGA DDDLDFTLPG LMPIEWQRFY NSRDERRDGL LGAGWSLPYE ISVQIEPHPE GGERLLYIDE QGR RIDMGS IALGAGAFSP GEGLSVRRNE NGQLLIESSD GIYRLFEPLP GDSSRLRLSL LGDRNDNRLH LEYDSSGRLW LLRD TFEQA QLELSYSTLW PGRIAQVERL YPDQQREVLV SYDYDAAGDL AQVRDSEGQV QRRFAYDAGH RMIEHQLATG LRCFY QWTQ FADQQWRVTR HWTDEGDSYQ YDYDLEAGIT QVTDSLQRLS RRRWNSQLQI VEYTDNLNQT WAFEWNDERQ LLSATD PLG GRYAYSYDET GNLIAETDPL GHSHSTLWLE HWSLPQTLTD PAGESWHLRY DPRGNCIAEV DPLGQVTQYR HDAHGQV VE IIDAAGRSRK MRWNSSGQLL EHLDCSGYPT RFEYDRRGNV KAIIDALGEH TRFHYDCQGR LLSSQLADGR AEHFQRSS N GQLQGYTDPS GYTTLYQYNR RGQIRQRIDA HGRQVRFDYD AYGRLLALTN ENGESYRFAW DAGNRLSEQQ DLDGSAKRY GYDLLNNVVE LQSHPAPYGT GLSAVPAQPI APIIHRFERD AVGRLLAKIT DDGRSQYTYD PLNQLTAASF TDNLGNQQAL SFSYDALGQ LLEEHTVAGS LIHRYDELGN LTQTQLPDKR WLNRLYYGSG HLHQINLDGQ VVSDFQRDRL HREVLRTQGQ I STRSEYDR CGRLRARLQH SSLLANTLPT APERHLEYDD SDNLVGLAER HATGQHRQLF HYDATGRIIA SQDGLQGQKE TF AYDAAAN LLDGPKAGAG LVVHNKLLTY QDKRYRYDAF GRMIEKRSGR RGVQRFAYDA EHRLIEVRNQ TSSGETLVRM RYD VLGRRI SKSEYDSQGN LLGETRFVWD GLRLLQEQRN QQTSLYVYED LGHAPLARVD GLGAQQKIRY YHNDLNGLPQ QLCE PDGHS VWQARYQVWG NTVEEIREPY YIEEQNLRFQ GQYLDRETGL HFNTFRFYDP DIGRFTTPDP IGLAGGLNLY QYAPN PIGW IDPLGWICKS AYSGRRGTTK AKADLERNGF TVVGEELTMK VKHPVTQKSY RIRADIIAKD KAGNYHVFEV KNGSGK LTK NQQGSGMFDM SNPANTNGGL GGGLIKPSGA TQGQFEVATK TARGTQFGGN GAQHSATFWL LRY UniProtKB: Rhs family protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 101.325 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
| Details | The sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 (4k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 1 / Number real images: 13090 / Average exposure time: 2.0 sec. / Average electron dose: 61.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 50 |
|---|---|
| Output model |  PDB-7q97: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)