+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12706 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Murine CIII2 focus-refined from supercomplex CICIII2 | |||||||||
 Map data Map data | murine CIII2 focus-refined from CICIII2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | mitochondria / respiratory chain / supercomplex / OXIDOREDUCTASE / complex III / complex IV / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationresponse to D-galactosamine / Complex III assembly / response to mercury ion / Respiratory electron transport / subthalamus development / pons development / response to cobalamin / cerebellar Purkinje cell layer development / mitochondrial respiratory chain complex III assembly / pyramidal neuron development ...response to D-galactosamine / Complex III assembly / response to mercury ion / Respiratory electron transport / subthalamus development / pons development / response to cobalamin / cerebellar Purkinje cell layer development / mitochondrial respiratory chain complex III assembly / pyramidal neuron development / response to alkaloid / Mitochondrial protein degradation / cellular respiration / thalamus development / respiratory chain complex III / quinol-cytochrome-c reductase / response to glucagon / quinol-cytochrome-c reductase activity / response to copper ion / mitochondrial electron transport, ubiquinol to cytochrome c / hypothalamus development / midbrain development / electron transport coupled proton transport / response to hyperoxia / animal organ regeneration / response to cadmium ion / response to hormone / response to activity / respiratory electron transport chain / hippocampus development / response to calcium ion / metalloendopeptidase activity / 2 iron, 2 sulfur cluster binding / response to toxic substance / myelin sheath / response to ethanol / response to hypoxia / mitochondrial inner membrane / response to xenobiotic stimulus / heme binding / ubiquitin protein ligase binding / protein-containing complex binding / mitochondrion / proteolysis / nucleoplasm / metal ion binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
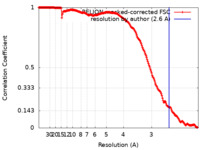
| Method | single particle reconstruction / cryo EM / Resolution: 2.6 Å | |||||||||
 Authors Authors | Vercellino I / Sazanov LA | |||||||||
| Funding support |  Austria, 1 items Austria, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Structure and assembly of the mammalian mitochondrial supercomplex CIIICIV. Authors: Irene Vercellino / Leonid A Sazanov /  Abstract: The enzymes of the mitochondrial electron transport chain are key players of cell metabolism. Despite being active when isolated, in vivo they associate into supercomplexes, whose precise role is ...The enzymes of the mitochondrial electron transport chain are key players of cell metabolism. Despite being active when isolated, in vivo they associate into supercomplexes, whose precise role is debated. Supercomplexes CIIICIV (refs. ), CICIII (ref. ) and CICIIICIV (respirasome) exist in mammals, but in contrast to CICIII and the respirasome, to date the only known eukaryotic structures of CIIICIV come from Saccharomyces cerevisiae and plants, which have different organization. Here we present the first, to our knowledge, structures of mammalian (mouse and ovine) CIIICIV and its assembly intermediates, in different conformations. We describe the assembly of CIIICIV from the CIII precursor to the final CIIICIV conformation, driven by the insertion of the N terminus of the assembly factor SCAF1 (ref. ) deep into CIII, while its C terminus is integrated into CIV. Our structures (which include CICIII and the respirasome) also confirm that SCAF1 is exclusively required for the assembly of CIIICIV and has no role in the assembly of the respirasome. We show that CIII is asymmetric due to the presence of only one copy of subunit 9, which straddles both monomers and prevents the attachment of a second copy of SCAF1 to CIII, explaining the presence of one copy of CIV in CIIICIV in mammals. Finally, we show that CIII and CIV gain catalytic advantage when assembled into the supercomplex and propose a role for CIIICIV in fine tuning the efficiency of electron transfer in the electron transport chain. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12706.map.gz emd_12706.map.gz | 71.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12706-v30.xml emd-12706-v30.xml emd-12706.xml emd-12706.xml | 28.8 KB 28.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12706_fsc.xml emd_12706_fsc.xml | 18.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_12706.png emd_12706.png | 156.1 KB | ||
| Filedesc metadata |  emd-12706.cif.gz emd-12706.cif.gz | 8.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12706 http://ftp.pdbj.org/pub/emdb/structures/EMD-12706 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12706 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12706 | HTTPS FTP |
-Validation report
| Summary document |  emd_12706_validation.pdf.gz emd_12706_validation.pdf.gz | 587.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12706_full_validation.pdf.gz emd_12706_full_validation.pdf.gz | 586.6 KB | Display | |
| Data in XML |  emd_12706_validation.xml.gz emd_12706_validation.xml.gz | 4.3 KB | Display | |
| Data in CIF |  emd_12706_validation.cif.gz emd_12706_validation.cif.gz | 4.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12706 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12706 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12706 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12706 | HTTPS FTP |
-Related structure data
| Related structure data |  7o3hMC  7o37C  7o3cC  7o3eC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12706.map.gz / Format: CCP4 / Size: 77.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12706.map.gz / Format: CCP4 / Size: 77.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | murine CIII2 focus-refined from CICIII2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
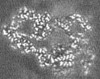
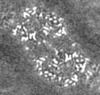
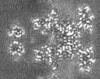
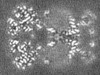
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.49981 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
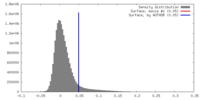
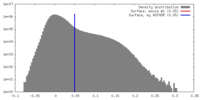
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Murine complex CIII2 focus-refined from supercomplex CICIII2
+Supramolecule #1: Murine complex CIII2 focus-refined from supercomplex CICIII2
+Macromolecule #1: Cytochrome b-c1 complex subunit 1, mitochondrial
+Macromolecule #2: Cytochrome b-c1 complex subunit 2, mitochondrial
+Macromolecule #3: Cytochrome b
+Macromolecule #4: Cytochrome c1, heme protein, mitochondrial
+Macromolecule #5: Cytochrome b-c1 complex subunit Rieske, mitochondrial
+Macromolecule #6: Cytochrome b-c1 complex subunit 7
+Macromolecule #7: Cytochrome b-c1 complex subunit 8
+Macromolecule #8: Cytochrome b-c1 complex subunit 6, mitochondrial
+Macromolecule #9: Cytochrome b-c1 complex subunit 9
+Macromolecule #10: Cytochrome b-c1 complex subunit 10
+Macromolecule #11: Cytochrome b-c1 complex subunit 9
+Macromolecule #12: 1,2-Distearoyl-sn-glycerophosphoethanolamine
+Macromolecule #13: CARDIOLIPIN
+Macromolecule #14: PROTOPORPHYRIN IX CONTAINING FE
+Macromolecule #15: HEME C
+Macromolecule #16: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #17: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.7 Component:
Details: CHAPS was added only upon freezing | |||||||||||||||
| Grid | Model: Quantifoil R0.6/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 1 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 5 sec. | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 7245 / Average exposure time: 1.17 sec. / Average electron dose: 90.66 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 2.7 µm / Calibrated defocus min: 0.2 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.4 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: OTHER / Overall B value: 65 / Target criteria: MAXIMAL LIKELIHOOD | ||||||
| Output model |  PDB-7o3h: |
 Movie
Movie Controller
Controller






















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)