[English] 日本語
 Yorodumi
Yorodumi- EMDB-12368: CryoEM structure of the human Separase-Cdk1-cyclin B1-Cks1 complex -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12368 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of the human Separase-Cdk1-cyclin B1-Cks1 complex | |||||||||
 Map data Map data | Separase-Cdk1-cyclin B1-Cks1 complex (postprocessed). | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | autoinhibition phosphate-binding pocket pseudosubstrate Scc1 / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of Schwann cell differentiation / cyclin A1-CDK1 complex / regulation of attachment of mitotic spindle microtubules to kinetochore / : / pronuclear fusion / response to DDT / negative regulation of sister chromatid cohesion / cyclin B1-CDK1 complex / separase / positive regulation of mitochondrial ATP synthesis coupled electron transport ...regulation of Schwann cell differentiation / cyclin A1-CDK1 complex / regulation of attachment of mitotic spindle microtubules to kinetochore / : / pronuclear fusion / response to DDT / negative regulation of sister chromatid cohesion / cyclin B1-CDK1 complex / separase / positive regulation of mitochondrial ATP synthesis coupled electron transport / regulation of chromosome condensation / Mitotic Prophase / positive regulation of mitotic sister chromatid segregation / histone kinase activity / meiotic chromosome separation / Golgi disassembly / E2F-enabled inhibition of pre-replication complex formation / microtubule cytoskeleton organization involved in mitosis / ventricular cardiac muscle cell development / Depolymerization of the Nuclear Lamina / MASTL Facilitates Mitotic Progression / positive regulation of mRNA 3'-end processing / regulation of mitotic cell cycle spindle assembly checkpoint / positive regulation of attachment of spindle microtubules to kinetochore / mitotic sister chromatid separation / Activation of NIMA Kinases NEK9, NEK6, NEK7 / patched binding / mitotic nuclear membrane disassembly / cyclin A2-CDK1 complex / Phosphorylation of Emi1 / homologous chromosome segregation / Transcriptional regulation by RUNX2 / establishment of mitotic spindle localization / Nuclear Pore Complex (NPC) Disassembly / Phosphorylation of the APC/C / G2/M DNA replication checkpoint / tissue regeneration / meiotic spindle organization / outer kinetochore / positive regulation of mitotic metaphase/anaphase transition / protein localization to kinetochore / Transcription of E2F targets under negative control by p107 (RBL1) and p130 (RBL2) in complex with HDAC1 / Initiation of Nuclear Envelope (NE) Reformation / Polo-like kinase mediated events / Golgi Cisternae Pericentriolar Stack Reorganization / cellular response to fatty acid / cyclin-dependent protein serine/threonine kinase activator activity / chromosome condensation / [RNA-polymerase]-subunit kinase / oocyte maturation / digestive tract development / response to amine / centrosome cycle / Condensation of Prometaphase Chromosomes / cyclin-dependent protein serine/threonine kinase regulator activity / response to copper ion / SCF ubiquitin ligase complex / mitotic metaphase chromosome alignment / cyclin-dependent protein kinase activity / MAPK3 (ERK1) activation / peptidyl-threonine phosphorylation / G1/S-Specific Transcription / mitotic G2 DNA damage checkpoint signaling / Regulation of APC/C activators between G1/S and early anaphase / ubiquitin-like protein ligase binding / microtubule organizing center / mitotic cytokinesis / mitotic sister chromatid segregation / catalytic activity / regulation of embryonic development / protein deubiquitination / positive regulation of G2/M transition of mitotic cell cycle / cysteine-type endopeptidase inhibitor activity / chromosome organization / cyclin-dependent kinase / response to mechanical stimulus / response to cadmium ion / cyclin-dependent protein serine/threonine kinase activity / response to axon injury / Regulation of MITF-M-dependent genes involved in cell cycle and proliferation / positive regulation of cardiac muscle cell proliferation / Cyclin A/B1/B2 associated events during G2/M transition / cyclin-dependent protein kinase holoenzyme complex / Chk1/Chk2(Cds1) mediated inactivation of Cyclin B:Cdk1 complex / Nuclear events stimulated by ALK signaling in cancer / ERK1 and ERK2 cascade / epithelial cell differentiation / Loss of Nlp from mitotic centrosomes / Loss of proteins required for interphase microtubule organization from the centrosome / RNA polymerase II CTD heptapeptide repeat kinase activity / cysteine-type peptidase activity / Recruitment of mitotic centrosome proteins and complexes / Hsp70 protein binding / positive regulation of mitotic cell cycle / Recruitment of NuMA to mitotic centrosomes / Anchoring of the basal body to the plasma membrane / APC/C:Cdc20 mediated degradation of Cyclin B / regulation of mitotic cell cycle / TP53 Regulates Transcription of Genes Involved in G2 Cell Cycle Arrest / molecular function activator activity Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Yu J / Raia P | |||||||||
| Funding support |  Switzerland, Switzerland,  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Structural basis of human separase regulation by securin and CDK1-cyclin B1. Authors: Jun Yu / Pierre Raia / Chloe M Ghent / Tobias Raisch / Yashar Sadian / Simone Cavadini / Pramod M Sabale / David Barford / Stefan Raunser / David O Morgan / Andreas Boland /     Abstract: In early mitosis, the duplicated chromosomes are held together by the ring-shaped cohesin complex. Separation of chromosomes during anaphase is triggered by separase-a large cysteine endopeptidase ...In early mitosis, the duplicated chromosomes are held together by the ring-shaped cohesin complex. Separation of chromosomes during anaphase is triggered by separase-a large cysteine endopeptidase that cleaves the cohesin subunit SCC1 (also known as RAD21). Separase is activated by degradation of its inhibitors, securin and cyclin B, but the molecular mechanisms of separase regulation are not clear. Here we used cryogenic electron microscopy to determine the structures of human separase in complex with either securin or CDK1-cyclin B1-CKS1. In both complexes, separase is inhibited by pseudosubstrate motifs that block substrate binding at the catalytic site and at nearby docking sites. As in Caenorhabditis elegans and yeast, human securin contains its own pseudosubstrate motifs. By contrast, CDK1-cyclin B1 inhibits separase by deploying pseudosubstrate motifs from intrinsically disordered loops in separase itself. One autoinhibitory loop is oriented by CDK1-cyclin B1 to block the catalytic sites of both separase and CDK1. Another autoinhibitory loop blocks substrate docking in a cleft adjacent to the separase catalytic site. A third separase loop contains a phosphoserine that promotes complex assembly by binding to a conserved phosphate-binding pocket in cyclin B1. Our study reveals the diverse array of mechanisms by which securin and CDK1-cyclin B1 bind and inhibit separase, providing the molecular basis for the robust control of chromosome segregation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12368.map.gz emd_12368.map.gz | 12.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12368-v30.xml emd-12368-v30.xml emd-12368.xml emd-12368.xml | 21.5 KB 21.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_12368.png emd_12368.png | 166.1 KB | ||
| Filedesc metadata |  emd-12368.cif.gz emd-12368.cif.gz | 8.2 KB | ||
| Others |  emd_12368_additional_1.map.gz emd_12368_additional_1.map.gz | 140.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12368 http://ftp.pdbj.org/pub/emdb/structures/EMD-12368 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12368 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12368 | HTTPS FTP |
-Related structure data
| Related structure data |  7nj0MC  7nj1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12368.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12368.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Separase-Cdk1-cyclin B1-Cks1 complex (postprocessed). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
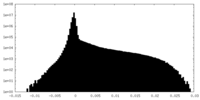
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Separase-Cdk1-cyclin B1-Cks1 complex (unsharpened).
| File | emd_12368_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Separase-Cdk1-cyclin B1-Cks1 complex (unsharpened). | ||||||||||||
| Projections & Slices |
| ||||||||||||
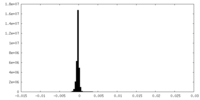
| Density Histograms |
- Sample components
Sample components
-Entire : Mutual inhibitory complex of human separase-Cdk1-cyclin B1-Cks1 (...
| Entire | Name: Mutual inhibitory complex of human separase-Cdk1-cyclin B1-Cks1 (CCC) complex. |
|---|---|
| Components |
|
-Supramolecule #1: Mutual inhibitory complex of human separase-Cdk1-cyclin B1-Cks1 (...
| Supramolecule | Name: Mutual inhibitory complex of human separase-Cdk1-cyclin B1-Cks1 (CCC) complex. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Securin,Separin
| Macromolecule | Name: Securin,Separin / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: separase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 244.677328 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQLGPPSPVK MPSPPWESNL LQSPSSILST LDVELPPVCC DIDIGGSGGS GGGSGGGSGE NLYFQGGGSG GSGMRSFKRV NFGTLLSSQ KEAEELLPDL KEFLSNPPAG FPSSRSDAER RQACDAILRA CNQQLTAKLA CPRHLGSLLE LAELACDGYL V STPQRPPL ...String: MQLGPPSPVK MPSPPWESNL LQSPSSILST LDVELPPVCC DIDIGGSGGS GGGSGGGSGE NLYFQGGGSG GSGMRSFKRV NFGTLLSSQ KEAEELLPDL KEFLSNPPAG FPSSRSDAER RQACDAILRA CNQQLTAKLA CPRHLGSLLE LAELACDGYL V STPQRPPL YLERILFVLL RNAAAQGSPE VTLRLAQPLH ACLVQCSREA APQDYEAVAR GSFSLLWKGA EALLERRAAF AA RLKALSF LVLLEDESTP CEVPHFASPT ACRAVAAHQL FDASGHGLNE ADADFLDDLL SRHVIRALVG ERGSSSGLLS PQR ALCLLE LTLEHCRRFC WSRHHDKAIS AVEKAHSYLR NTNLAPSLQL CQLGVKLLQV GEEGPQAVAK LLIKASAVLS KSME APSPP LRALYESCQF FLSGLERGTK RRYRLDAILS LFAFLGGYCS LLQQLRDDGV YGGSSKQQQS FLQMYFQGLH LYTVV VYDF AQGCQIVDLA DLTQLVDSCK STVVWMLEAL EGLSGQELTD HMGMTASYTS NLAYSFYSHK LYAEACAISE PLCQHL GLV KPGTYPEVPP EKLHRCFRLQ VESLKKLGKQ AQGCKMVILW LAALQPCSPE HMAEPVTFWV RVKMDAARAG DKELQLK TL RDSLSGWDPE TLALLLREEL QAYKAVRADT GQERFNIICD LLELSPEETP AGAWARATHL VELAQVLCYH DFTQQTNC S ALDAIREALQ LLDSVRPEAQ ARDQLLDDKA QALLWLYICT LEAKIQEGIE RDRRAQAPGN LEEFEVNDLN YEDKLQEDR FLYSNIAFNL AADAAQSKCL DQALALWKEL LTKGQAPAVR CLQQTAASLQ ILAALYQLVA KPMQALEVLL LLRIVSERLK DHSKAAGSS CHITQLLLTL GCPSYAQLHL EEAASSLKHL DQTTDTYLLL SLTCDLLRSQ LYWTHQKVTK GVSLLLSVLR D PALQKSSK AWYLLRVQVL QLVAAYLSLP SNNLSHSLWE QLCAQGWQTP EIALIDSHKL LRSIILLLMG SDILSTQKAA VE TSFLDYG ENLVQKWQVL SEVLSCSEKL VCHLGRLGSV SEAKAFCLEA LKLTTKLQIP RQCALFLVLK GELELARNDI DLC QSDLQQ VLFLLESCTE FGGVTQHLDS VKKVHLQKGK QQAQVPCPPQ LPEEELFLRG PALELVATVA KEPGPIAPST NS (SEP)PVLKTK PQPIPNFLSH SPTCDCSLCA SPVLTAVCLR WVLVTAGVRL AMGHQAQGLD LLQVVLKGCP EAAERLTQA LQASLNHKTP PSLVPSLLDE ILAQAYTLLA LEGLNQPSNE SLQKVLQSGL KFVAARIPHL EPWRASLLLI WALTKLGGLS CCTTQLFAS SWGWQPPLIK SVPGSEPSKT QGQKRSGRGR QKLASAPLSL NNTSQKGLEG RGLPCTPKPP DRIRQAGPHV P FTVFEEVC PTESKPEVPQ APRVQQRVQT RLKVNFSDDS DLEDPVSAEA WLAEEPKRRG TASRGRGRAR KGLSLKTDAV VA PGSAPGN PGLNGRSRRA KKVASRHCEE RRPQRASDQA RPGPEIMRTI PEEELTDNWR KMSFEILRGS DGEDSASGGK TPA PGPEAA SGEWELLRLD SSKKKLPSPC PDKESDKDLG PRLQLPSAPV ATGLSTLDSI CDSLSVAFRG ISHCPPSGLY AHLC RFLAL CLGHRDPYAT AFLVTESVSI TCRHQLLTHL HRQLSKAQKH RGSLEIADQL QGLSLQEMPG DVPLARIQRL FSFRA LESG HFPQPEKESF QERLALIPSG VTVCVLALAT LQPGTVGNTL LLTRLEKDSP PVSVQIPTGQ NKLHLRSVLN EFDAIQ KAQ KENSSCTDKR EWWTGRLALD HRMEVLIASL EKSVLGCWKG LLLPSSEEPG PAQEASRLQE LLQDCGWKYP DRTLLKI ML SGAGALTPQD IQALAYGLCP TQPERAQELL NEAVGRLQGL TVPSNSHLVL VLDKDLQKLP WESMPSLQAL PVTRLPSF R FLLSYSIIKE YGASPVLSQG VDPRSTFYVL NPHNNLSSTE EQFRANFSSE AGWRGVVGEV PRPEQVQEAL TKHDLYIYA GHGAGARFLD GQAVLRLSCR AVALLFGSSS AALAVHGNLE GAGIVLKYIM AGCPLFLGNL WDVTDRDIDR YTEALLQGWL GAGPGAPLL YYVNQARQAP RLKYLIGAAP IAYGLPVSLR SSLAEENLYF QSWSHPQFEK GGGSGGGSGG GSWSHPQFEK UniProtKB: Securin, Separin |
-Macromolecule #2: Cyclin-dependent kinase 1
| Macromolecule | Name: Cyclin-dependent kinase 1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: cyclin-dependent kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 36.667098 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEDYTKIEKI GEGTYGVVYK GRHKTTGQVV AMKKIRLESE EEGVPSTAIR EISLLKELRH PNIVSLQDVL MQDSRLYLIF EFLSMDLKK YLDSIPPGQY MDSSLVKSYL YQILQGIVFC HSRRVLHRDL KPQNLLIDDK GTIKLADFGL ARAFGIPIRV Y (TPO)HEVVTLW ...String: MEDYTKIEKI GEGTYGVVYK GRHKTTGQVV AMKKIRLESE EEGVPSTAIR EISLLKELRH PNIVSLQDVL MQDSRLYLIF EFLSMDLKK YLDSIPPGQY MDSSLVKSYL YQILQGIVFC HSRRVLHRDL KPQNLLIDDK GTIKLADFGL ARAFGIPIRV Y (TPO)HEVVTLW YRSPEVLLGS ARYSTPVDIW SIGTIFAELA TKKPLFHGDS EIDQLFRIFR ALGTPNNEVW PEVESLQD Y KNTFPKWKPG SLASHVKNLD ENGLDLLSKM LIYDPAKRIS GKMALNHPYF NDLDNQIKKM IAAEALEVLF QGPHHHHHH HH UniProtKB: Cyclin-dependent kinase 1 |
-Macromolecule #3: G2/mitotic-specific cyclin-B1,G2/mitotic-specific cyclin-B1
| Macromolecule | Name: G2/mitotic-specific cyclin-B1,G2/mitotic-specific cyclin-B1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 52.625723 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MALRVTRNSK INAENKAKIN MAGAKRVPTA PAATSKPGLR PRTALGDIGN KVSEQLQAKM PMKKEAKPSA TGKVIDKKLP KPLEKVPML VPVPVSEPVP EPEPEPEPEP VKEEKLSPEP ILVDTASPSP METSGCAPAE EDLCQAFSDV ILAVNDVDAE D GADPNLCS ...String: MALRVTRNSK INAENKAKIN MAGAKRVPTA PAATSKPGLR PRTALGDIGN KVSEQLQAKM PMKKEAKPSA TGKVIDKKLP KPLEKVPML VPVPVSEPVP EPEPEPEPEP VKEEKLSPEP ILVDTASPSP METSGCAPAE EDLCQAFSDV ILAVNDVDAE D GADPNLCS EYVKDIYAYL RQLEEEQAVR PKYLLGREVT GNMRAILIDW LVQVQMKFRL LQETMYMTVS IIDRFMQNNC VP KKMLQLV GVTAMFIASK YEEMYPPEIG DFAFVTDNTY TKHQIRQMEM KILRALNFGL GRPLPLHFLR RASKIGEVDV EQH TLAKYL MELTMLDYDM VHFPPSQIAA GAFCLALKIL DNGEWTPTLQ HYLSYTEESL LPVMQHLAKN VVMVNQGLTK HMTV KNKYA TSKHAKISTL PQLNSALVQD LAKAVAKVSS LAEENLYFQS WSHPQFEKGG GSGGGSGGGS WSHPQFEK UniProtKB: G2/mitotic-specific cyclin-B1 |
-Macromolecule #4: Cyclin-dependent kinases regulatory subunit 1
| Macromolecule | Name: Cyclin-dependent kinases regulatory subunit 1 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 9.679211 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSHKQIYYSD KYDDEEFEYR HVMLPKDIAK LVPKTHLMSE SEWRNLGVQQ SQGWVHYMIH EPEPHILLFR RPLPKKPKK UniProtKB: Cyclin-dependent kinases regulatory subunit 1 |
-Macromolecule #5: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.050 mg/mL |
|---|---|
| Buffer | pH: 7.8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Instrument: LEICA EM GP |
| Details | The sample was monodisperse. We use graphene oxide-coated EM grids. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 5 / Number real images: 13640 / Average exposure time: 3.0 sec. / Average electron dose: 78.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 2.5 µm / Calibrated defocus min: 1.3 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.3 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: RELION / Number images used: 312836 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)