+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13843 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of VgrG1 from Pseudomonas protegens. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacterial type VI secretion system / TOXIN / VgrG | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Pseudomonas protegens Pf-5 (bacteria) / Pseudomonas protegens Pf-5 (bacteria) /  Pseudomonas fluorescens (strain ATCC BAA-477 / NRRL B-23932 / Pf-5) (bacteria) Pseudomonas fluorescens (strain ATCC BAA-477 / NRRL B-23932 / Pf-5) (bacteria) | |||||||||
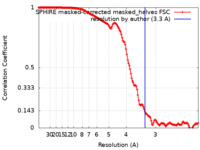
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Guenther P / Quentin D | |||||||||
| Funding support | 2 items
| |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2022 Journal: PLoS Pathog / Year: 2022Title: Structure of a bacterial Rhs effector exported by the type VI secretion system. Authors: Patrick Günther / Dennis Quentin / Shehryar Ahmad / Kartik Sachar / Christos Gatsogiannis / John C Whitney / Stefan Raunser /   Abstract: The type VI secretion system (T6SS) is a widespread protein export apparatus found in Gram-negative bacteria. The majority of T6SSs deliver toxic effector proteins into competitor bacteria. Yet, the ...The type VI secretion system (T6SS) is a widespread protein export apparatus found in Gram-negative bacteria. The majority of T6SSs deliver toxic effector proteins into competitor bacteria. Yet, the structure, function, and activation of many of these effectors remains poorly understood. Here, we present the structures of the T6SS effector RhsA from Pseudomonas protegens and its cognate T6SS spike protein, VgrG1, at 3.3 Å resolution. The structures reveal that the rearrangement hotspot (Rhs) repeats of RhsA assemble into a closed anticlockwise β-barrel spiral similar to that found in bacterial insecticidal Tc toxins and in metazoan teneurin proteins. We find that the C-terminal toxin domain of RhsA is autoproteolytically cleaved but remains inside the Rhs 'cocoon' where, with the exception of three ordered structural elements, most of the toxin is disordered. The N-terminal 'plug' domain is unique to T6SS Rhs proteins and resembles a champagne cork that seals the Rhs cocoon at one end while also mediating interactions with VgrG1. Interestingly, this domain is also autoproteolytically cleaved inside the cocoon but remains associated with it. We propose that mechanical force is required to remove the cleaved part of the plug, resulting in the release of the toxin domain as it is delivered into a susceptible bacterial cell by the T6SS. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13843.map.gz emd_13843.map.gz | 2.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13843-v30.xml emd-13843-v30.xml emd-13843.xml emd-13843.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13843_fsc.xml emd_13843_fsc.xml | 18.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_13843.png emd_13843.png | 27.7 KB | ||
| Masks |  emd_13843_msk_1.map emd_13843_msk_1.map | 259.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13843.cif.gz emd-13843.cif.gz | 6.3 KB | ||
| Others |  emd_13843_half_map_1.map.gz emd_13843_half_map_1.map.gz emd_13843_half_map_2.map.gz emd_13843_half_map_2.map.gz | 125.2 MB 125.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13843 http://ftp.pdbj.org/pub/emdb/structures/EMD-13843 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13843 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13843 | HTTPS FTP |
-Related structure data
| Related structure data |  7q5pMC  7q97C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13843.map.gz / Format: CCP4 / Size: 259.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13843.map.gz / Format: CCP4 / Size: 259.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13843_msk_1.map emd_13843_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_13843_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_13843_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of the VgrG1 trimer from Pseudomonas protegens.
| Entire | Name: Structure of the VgrG1 trimer from Pseudomonas protegens. |
|---|---|
| Components |
|
-Supramolecule #1: Structure of the VgrG1 trimer from Pseudomonas protegens.
| Supramolecule | Name: Structure of the VgrG1 trimer from Pseudomonas protegens. type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Pseudomonas protegens Pf-5 (bacteria) Pseudomonas protegens Pf-5 (bacteria) |
| Molecular weight | Theoretical: 215.994 KDa |
-Macromolecule #1: Type VI secretion protein VgrG
| Macromolecule | Name: Type VI secretion protein VgrG / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas fluorescens (strain ATCC BAA-477 / NRRL B-23932 / Pf-5) (bacteria) Pseudomonas fluorescens (strain ATCC BAA-477 / NRRL B-23932 / Pf-5) (bacteria)Strain: ATCC BAA-477 / NRRL B-23932 / Pf-5 |
| Molecular weight | Theoretical: 72.080133 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLFQQSTRLA QVNCPLGPDV LLLKSLGGGE ELGRLFDYQL QLASSDANID LNQLLGKPMG LSVQLDGGGQ RYFHGIVARC SQNIDTGQF ASYEVTLRPW LWLLSRTSDC RIFQNLSIPQ IIKQVFRDLG FSDFEDALSR PYREWEYCVQ YRETSFDFVS R LMEQEGIY ...String: MLFQQSTRLA QVNCPLGPDV LLLKSLGGGE ELGRLFDYQL QLASSDANID LNQLLGKPMG LSVQLDGGGQ RYFHGIVARC SQNIDTGQF ASYEVTLRPW LWLLSRTSDC RIFQNLSIPQ IIKQVFRDLG FSDFEDALSR PYREWEYCVQ YRETSFDFVS R LMEQEGIY YFFRHEKDRH VVVLADAYGA HSSVPGYASV PYYPRDEQQR ERDHMFDWHL AQEVQPGSLE LNDYDFQRPS AR IDVRSAM PRPHSAGDYP LYDYPGTYVQ SSDGEHYAQT RIEALQSLHE RIELSGNARG LGVGNLFSLT GFSRQDQNRE YLI VSIRYY LVQESLESGA GGGSAQFESH LTCIDAQQSF RPLATTHKPM VQGPQTARVV GPAGEEIWTD QYGRVKVHFH WDRH DQSNE NSSCWIRVSQ AWAGKNWGSM QIPRIGQEVI VSFLEGDPDR PIITGRVYNA EQTVPYDLPA NATQSGMKSR SSKGG SPAN FNEIRMEDKK GAEQLYIHAE RNQDIVVEVN ESHSVGNNRN KSIGHDEYVT IGNKRTRIVQ HVDELRVGEK KLDSVG QSY VIEVGERLRL VCGASILELN ASGQINLCGV NISVHASADA QINTGGVLHL NNGGGPGTTT EGQGVQGAIS AKAKAPF SA PKG UniProtKB: Type VI secretion protein VgrG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 101.325 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV |
| Details | The sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Specialist optics | Spherical aberration corrector: Cs corrector / Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 1250 / Average exposure time: 1.5 sec. / Average electron dose: 90.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)