[English] 日本語
 Yorodumi
Yorodumi- EMDB-13424: Structure of Multidrug and Toxin Compound Extrusion (MATE) transp... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13424 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Multidrug and Toxin Compound Extrusion (MATE) transporter NorM by NabFab-fiducial assisted cryo-EM | |||||||||
 Map data Map data | Map with fulcrum ion center of chimeric nanobody | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NabFab / anti-nanobody Fab / fiducial / NorM / MATE / MEMBRANE PROTEIN | |||||||||
| Function / homology | Multi antimicrobial extrusion protein / MatE / antiporter activity / xenobiotic transmembrane transporter activity / membrane => GO:0016020 / Multidrug resistance protein NorM Function and homology information Function and homology information | |||||||||
| Biological species |  Vibrio cholerae RC385 (bacteria) / synthetic construct (others) / Vibrio cholerae RC385 (bacteria) / synthetic construct (others) /  | |||||||||
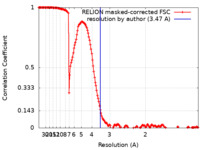
| Method | single particle reconstruction / cryo EM / Resolution: 3.47 Å | |||||||||
 Authors Authors | Bloch JS / Mukherjee S | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins. Authors: Joël S Bloch / Somnath Mukherjee / Julia Kowal / Ekaterina V Filippova / Martina Niederer / Els Pardon / Jan Steyaert / Anthony A Kossiakoff / Kaspar P Locher /    Abstract: With conformation-specific nanobodies being used for a wide range of structural, biochemical, and cell biological applications, there is a demand for antigen-binding fragments (Fabs) that ...With conformation-specific nanobodies being used for a wide range of structural, biochemical, and cell biological applications, there is a demand for antigen-binding fragments (Fabs) that specifically and tightly bind these nanobodies without disturbing the nanobody-target protein interaction. Here, we describe the development of a synthetic Fab (termed NabFab) that binds the scaffold of an alpaca-derived nanobody with picomolar affinity. We demonstrate that upon complementary-determining region grafting onto this parent nanobody scaffold, nanobodies recognizing diverse target proteins and derived from llama or camel can cross-react with NabFab without loss of affinity. Using NabFab as a fiducial and size enhancer (50 kDa), we determined the high-resolution cryogenic electron microscopy (cryo-EM) structures of nanobody-bound VcNorM and ScaDMT, both small membrane proteins of ∼50 kDa. Using an additional anti-Fab nanobody further facilitated reliable initial three-dimensional structure determination from small cryo-EM test datasets. Given that NabFab is of synthetic origin, is humanized, and can be conveniently expressed in in large amounts, it may be useful not only for structural biology but also for biomedical applications. #1:  Journal: Biorxiv / Year: 2021 Journal: Biorxiv / Year: 2021Title: Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins Authors: Bloch JS / Mukherjee S / Kowal J / Filippova EV / Niederer M / Pardon E / Steyaert J / Kossiakoff AA / Locher KP | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13424.map.gz emd_13424.map.gz | 320.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13424-v30.xml emd-13424-v30.xml emd-13424.xml emd-13424.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13424_fsc.xml emd_13424_fsc.xml | 16.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_13424.png emd_13424.png | 67.7 KB | ||
| Filedesc metadata |  emd-13424.cif.gz emd-13424.cif.gz | 6.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13424 http://ftp.pdbj.org/pub/emdb/structures/EMD-13424 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13424 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13424 | HTTPS FTP |
-Validation report
| Summary document |  emd_13424_validation.pdf.gz emd_13424_validation.pdf.gz | 542.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_13424_full_validation.pdf.gz emd_13424_full_validation.pdf.gz | 541.8 KB | Display | |
| Data in XML |  emd_13424_validation.xml.gz emd_13424_validation.xml.gz | 14.3 KB | Display | |
| Data in CIF |  emd_13424_validation.cif.gz emd_13424_validation.cif.gz | 19.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13424 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13424 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13424 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-13424 | HTTPS FTP |
-Related structure data
| Related structure data |  7phpMC  7phqC  7pijC  7rthC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13424.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13424.map.gz / Format: CCP4 / Size: 347.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map with fulcrum ion center of chimeric nanobody | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.67 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex of vibrio cholerae MATE transporter NorM, chimeric nanobo...
| Entire | Name: Complex of vibrio cholerae MATE transporter NorM, chimeric nanobody NorM-Nb17_4, NabFab, and anti-Fab nanobody |
|---|---|
| Components |
|
-Supramolecule #1: Complex of vibrio cholerae MATE transporter NorM, chimeric nanobo...
| Supramolecule | Name: Complex of vibrio cholerae MATE transporter NorM, chimeric nanobody NorM-Nb17_4, NabFab, and anti-Fab nanobody type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Vibrio cholerae RC385 (bacteria) Vibrio cholerae RC385 (bacteria) |
-Macromolecule #1: Multidrug resistance protein NorM
| Macromolecule | Name: Multidrug resistance protein NorM / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio cholerae RC385 (bacteria) Vibrio cholerae RC385 (bacteria) |
| Molecular weight | Theoretical: 52.462109 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHHH HHSSGLEVLF QGPSGTSVHR YKKEASNLIK LATPVLIASV AQTGMGFVDT IMAGGVSAID MAAVSIAASI WLPSILFGV GLLMALVPVV AQLNGAGRQH KIPFEVHQGL ILALLVSIPI IAVLFQTQFI IRFMDVEEAM ATKTVGYMHA V IFAVPAYL ...String: MGHHHHHHHH HHSSGLEVLF QGPSGTSVHR YKKEASNLIK LATPVLIASV AQTGMGFVDT IMAGGVSAID MAAVSIAASI WLPSILFGV GLLMALVPVV AQLNGAGRQH KIPFEVHQGL ILALLVSIPI IAVLFQTQFI IRFMDVEEAM ATKTVGYMHA V IFAVPAYL LFQALRSFTD GMSLTKPAMV IGFIGLLLNI PLNWIFVYGK FGAPELGGVG CGVATAIVYW IMLLLLLFYI VT SKRLAHV KVFETFHKPQ PKELIRLFRL GFPVAAALFF EVTLFAVVAL LVAPLGSTVV AAHQVALNFS SLVFMFPMSI GAA VSIRVG HKLGEQDTKG AAIAANVGLM TGLATACITA LLTVLFREQI ALLYTENQVV VALAMQLLLF AAIYQCMDAV QVVA AGSLR GYKDMTAIFH RTFISYWVLG LPTGYILGMT NWLTEQPLGA KGFWLGFIIG LSAAALMLGQ RLYWLQKQSD DVQLH LAAK UniProtKB: Multidrug resistance protein NorM |
-Macromolecule #2: NabFab HC
| Macromolecule | Name: NabFab HC / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 25.684463 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NFSYYSIHWV RQAPGKGLEW VAYISSSSSY TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARGYQYWQYH ASWYWNGGLD YWGQGTLVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL V KDYFPEPV ...String: EISEVQLVES GGGLVQPGGS LRLSCAASGF NFSYYSIHWV RQAPGKGLEW VAYISSSSSY TSYADSVKGR FTISADTSKN TAYLQMNSL RAEDTAVYYC ARGYQYWQYH ASWYWNGGLD YWGQGTLVTV SSASTKGPSV FPLAPSSKST SGGTAALGCL V KDYFPEPV TVSWNSGALT SGVHTFPAVL QSSGLYSLSS VVTVPSSSLG TQTYICNVNH KPSNTKVDKK VEPKSCDKTH T |
-Macromolecule #3: NabFab LC
| Macromolecule | Name: NabFab LC / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 23.258783 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSSSSLITF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: SDIQMTQSPS SLSASVGDRV TITCRASQSV SSAVAWYQQK PGKAPKLLIY SASSLYSGVP SRFSGSRSGT DFTLTISSLQ PEDFATYYC QQSSSSLITF GQGTKVEIKR TVAAPSVFIF PPSDSQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #4: NorM-Nb17_4
| Macromolecule | Name: NorM-Nb17_4 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 14.356979 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: QRQLVESGGG LVQPGGSLRL SCAASGIIFK INDMGWFRQA PGKEREGVAG ITSGGRTNYA DSVKGRFIIS RDNVKNTVYL QMNSLEPED TAVYYCKSDG LISYAASQLS TYWGKGTPVT VSSHHHHHHE PEA |
-Macromolecule #5: Anti-Fab nanobody
| Macromolecule | Name: Anti-Fab nanobody / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.390644 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSQVQLQESG GGLVQPGGSL RLSCAASGRT ISRYAMSWFR QAPGKEREFV AVARRSGDGA FYADSVQGRF TVSRDDAKNT VYLQMNSLK PEDTAVYYCA IDSDTFYSGS YDYWGQGTQV TVSS |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 21 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 26.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)