[English] 日本語
 Yorodumi
Yorodumi- EMDB-13271: Subtomogram averaging of apoferritin at 2.86 angstrom resolution -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13271 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Subtomogram averaging of apoferritin at 2.86 angstrom resolution | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
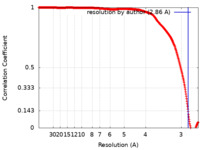
| Method | subtomogram averaging / cryo EM / Resolution: 2.86 Å | |||||||||
 Authors Authors | Ni T / Mendonca L / Frosio T / Sheng Y / Clare D / Himes B / Zhang P | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Protoc / Year: 2022 Journal: Nat Protoc / Year: 2022Title: High-resolution in situ structure determination by cryo-electron tomography and subtomogram averaging using emClarity. Authors: Tao Ni / Thomas Frosio / Luiza Mendonça / Yuewen Sheng / Daniel Clare / Benjamin A Himes / Peijun Zhang /   Abstract: Cryo-electron tomography and subtomogram averaging (STA) has developed rapidly in recent years. It provides structures of macromolecular complexes in situ and in cellular context at or below ...Cryo-electron tomography and subtomogram averaging (STA) has developed rapidly in recent years. It provides structures of macromolecular complexes in situ and in cellular context at or below subnanometer resolution and has led to unprecedented insights into the inner working of molecular machines in their native environment, as well as their functional relevant conformations and spatial distribution within biological cells or tissues. Given the tremendous potential of cryo-electron tomography STA in in situ structural cell biology, we previously developed emClarity, a graphics processing unit-accelerated image-processing software that offers STA and classification of macromolecular complexes at high resolution. However, the workflow remains challenging, especially for newcomers to the field. In this protocol, we describe a detailed workflow, processing and parameters associated with each step, from initial tomography tilt-series data to the final 3D density map, with several features unique to emClarity. We use four different samples, including human immunodeficiency virus type 1 Gag assemblies, ribosome and apoferritin, to illustrate the procedure and results of STA and classification. Following the processing steps described in this protocol, along with a comprehensive tutorial and guidelines for troubleshooting and parameter optimization, one can obtain density maps up to 2.8 Å resolution from six tilt series by cryo-electron tomography STA. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13271.map.gz emd_13271.map.gz | 3.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13271-v30.xml emd-13271-v30.xml emd-13271.xml emd-13271.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13271_fsc.xml emd_13271_fsc.xml | 34.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_13271.png emd_13271.png | 153.7 KB | ||
| Others |  emd_13271_half_map_1.map.gz emd_13271_half_map_1.map.gz emd_13271_half_map_2.map.gz emd_13271_half_map_2.map.gz | 3 MB 2.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13271 http://ftp.pdbj.org/pub/emdb/structures/EMD-13271 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13271 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13271 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10787 (Title: cryo electron tomography of Apoferritin / Data size: 17.3 EMPIAR-10787 (Title: cryo electron tomography of Apoferritin / Data size: 17.3 Data #1: Unaligned raw tilt-series of apoferritin on graphene grid [tilt series]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13271.map.gz / Format: CCP4 / Size: 21.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13271.map.gz / Format: CCP4 / Size: 21.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
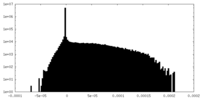
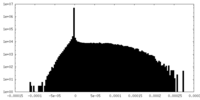
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #2
| File | emd_13271_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_13271_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : apoferritin
| Entire | Name: apoferritin |
|---|---|
| Components |
|
-Supramolecule #1: apoferritin
| Supramolecule | Name: apoferritin / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: GRAPHENE / Support film - topology: CONTINUOUS / Support film - Film thickness: 0.8 nm / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Average electron dose: 102.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)