[English] 日本語
 Yorodumi
Yorodumi- EMDB-13089: major seeded in vitro fibril morphology from murine SAA1.1 protein -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-13089 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | major seeded in vitro fibril morphology from murine SAA1.1 protein | |||||||||||||||||||||
 Map data Map data | Masked post-processed density map of the reconstructed murine SAA1.1 fibril | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | systemic amyloidosis / seeded / misfolding disease / inflammation / prion / protein fibril | |||||||||||||||||||||
| Function / homology | Serum amyloid A protein / : / Serum amyloid A protein / Serum amyloid A proteins signature. / Serum amyloid A proteins / response to stilbenoid / high-density lipoprotein particle / acute-phase response / Serum amyloid A-2 protein Function and homology information Function and homology information | |||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||
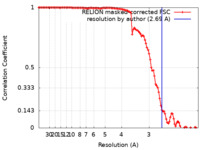
| Method | helical reconstruction / cryo EM / Resolution: 2.69 Å | |||||||||||||||||||||
 Authors Authors | Heerde T / Schmidt M | |||||||||||||||||||||
| Funding support |  Germany, 6 items Germany, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Cryo-EM demonstrates the in vitro proliferation of an ex vivo amyloid fibril morphology by seeding. Authors: Thomas Heerde / Matthies Rennegarbe / Alexander Biedermann / Dilan Savran / Peter B Pfeiffer / Manuel Hitzenberger / Julian Baur / Ioana Puscalau-Girtu / Martin Zacharias / Nadine Schwierz / ...Authors: Thomas Heerde / Matthies Rennegarbe / Alexander Biedermann / Dilan Savran / Peter B Pfeiffer / Manuel Hitzenberger / Julian Baur / Ioana Puscalau-Girtu / Martin Zacharias / Nadine Schwierz / Christian Haupt / Matthias Schmidt / Marcus Fändrich /  Abstract: Several studies showed that seeding of solutions of monomeric fibril proteins with ex vivo amyloid fibrils accelerated the kinetics of fibril formation in vitro but did not necessarily replicate the ...Several studies showed that seeding of solutions of monomeric fibril proteins with ex vivo amyloid fibrils accelerated the kinetics of fibril formation in vitro but did not necessarily replicate the seed structure. In this research we use cryo-electron microscopy and other methods to analyze the ability of serum amyloid A (SAA)1.1-derived amyloid fibrils, purified from systemic AA amyloidosis tissue, to seed solutions of recombinant SAA1.1 protein. We show that 98% of the seeded fibrils remodel the full fibril structure of the main ex vivo fibril morphology, which we used for seeding, while they are notably different from unseeded in vitro fibrils. The seeded fibrils show a similar proteinase K resistance as ex vivo fibrils and are substantially more stable to proteolytic digestion than unseeded in vitro fibrils. Our data support the view that the fibril morphology contributes to determining proteolytic stability and that pathogenic amyloid fibrils arise from proteolytic selection. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13089.map.gz emd_13089.map.gz | 5.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13089-v30.xml emd-13089-v30.xml emd-13089.xml emd-13089.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13089_fsc.xml emd_13089_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_13089.png emd_13089.png | 51.9 KB | ||
| Masks |  emd_13089_msk_1.map emd_13089_msk_1.map | 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13089.cif.gz emd-13089.cif.gz | 5.9 KB | ||
| Others |  emd_13089_half_map_1.map.gz emd_13089_half_map_1.map.gz emd_13089_half_map_2.map.gz emd_13089_half_map_2.map.gz | 80.4 MB 80.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13089 http://ftp.pdbj.org/pub/emdb/structures/EMD-13089 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13089 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13089 | HTTPS FTP |
-Related structure data
| Related structure data |  7ovtMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13089.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13089.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Masked post-processed density map of the reconstructed murine SAA1.1 fibril | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
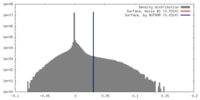
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_13089_msk_1.map emd_13089_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
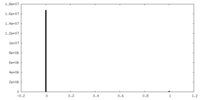
| Density Histograms |
-Half map: Second half map of the reconstructed fibril
| File | emd_13089_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Second half map of the reconstructed fibril | ||||||||||||
| Projections & Slices |
| ||||||||||||
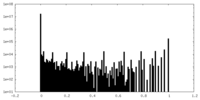
| Density Histograms |
-Half map: First half map of the reconstructed fibril
| File | emd_13089_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | First half map of the reconstructed fibril | ||||||||||||
| Projections & Slices |
| ||||||||||||
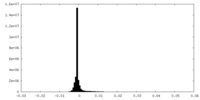
| Density Histograms |
- Sample components
Sample components
-Entire : Murine serum amyloid A1 (SAA1) amyloid fibril
| Entire | Name: Murine serum amyloid A1 (SAA1) amyloid fibril |
|---|---|
| Components |
|
-Supramolecule #1: Murine serum amyloid A1 (SAA1) amyloid fibril
| Supramolecule | Name: Murine serum amyloid A1 (SAA1) amyloid fibril / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: in vitro murine SAA amyloid fibril morphology i; Seeded with ex vivo material |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Serum amyloid A-2 protein
| Macromolecule | Name: Serum amyloid A-2 protein / type: protein_or_peptide / ID: 1 / Details: amyloid fibril / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.622629 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GFFSFIGEAF QGAGDMWRAY TDMKEAGWKD GDKYFHARGN YDAAQRGPGG VWAAEKISDA RESFQEFFGR GHEDTMADQE ANRHGRSGK DPNYYRPPGL PAKY UniProtKB: Serum amyloid A-2 protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 8.5 / Component - Concentration: 10.0 mM / Component - Formula: (HOCH2)3CNH2 / Component - Name: Tris(hydroxymethyl)aminomethane / Details: 10mM Tris(hydroxymethyl)aminomethane (Tris) |
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 96 % / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average exposure time: 12.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: BACKBONE TRACE / Target criteria: correlation coefficient |
|---|---|
| Output model |  PDB-7ovt: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)