[English] 日本語
 Yorodumi
Yorodumi- EMDB-12962: Structure of the outer-membrane core complex (inner ring) from a ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12962 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the outer-membrane core complex (inner ring) from a conjugative type IV secretion system | |||||||||
 Map data Map data | Structure of the outer-membrane core complex (inner ring) from a conjugative type IV secretion system | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Type IV secretion system / F plasmid / outer-membrane core complex / conjugation / MEMBRANE PROTEIN | |||||||||
| Function / homology | Type IV conjugative transfer system protein TraV / Type IV conjugative transfer system lipoprotein (TraV) / Prokaryotic membrane lipoprotein lipid attachment site profile. / Type IV conjugative transfer system lipoprotein TraV Function and homology information Function and homology information | |||||||||
| Biological species |  Salmonella enterica (bacteria) Salmonella enterica (bacteria) | |||||||||
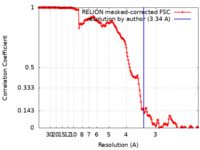
| Method | single particle reconstruction / cryo EM / Resolution: 3.34 Å | |||||||||
 Authors Authors | Amin H / Ilangovan A / Costa TRD | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Architecture of the outer-membrane core complex from a conjugative type IV secretion system. Authors: Himani Amin / Aravindan Ilangovan / Tiago R D Costa /  Abstract: Conjugation is one of the most important processes that bacteria utilize to spread antibiotic resistance genes among bacterial populations. Interbacterial DNA transfer requires a large double ...Conjugation is one of the most important processes that bacteria utilize to spread antibiotic resistance genes among bacterial populations. Interbacterial DNA transfer requires a large double membrane-spanning nanomachine called the type 4 secretion system (T4SS) made up of the inner-membrane complex (IMC), the outer-membrane core complex (OMCC) and the conjugative pilus. The iconic F plasmid-encoded T4SS has been central in understanding conjugation for several decades, however atomic details of its structure are not known. Here, we report the structure of a complete conjugative OMCC encoded by the pED208 plasmid from E. coli, solved by cryo-electron microscopy at 3.3 Å resolution. This 2.1 MDa complex has a unique arrangement with two radial concentric rings, each having a different symmetry eventually contributing to remarkable differences in protein stoichiometry and flexibility in comparison to other OMCCs. Our structure suggests that F-OMCC is a highly dynamic complex, with implications for pilus extension and retraction during conjugation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12962.map.gz emd_12962.map.gz | 392 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12962-v30.xml emd-12962-v30.xml emd-12962.xml emd-12962.xml | 10.4 KB 10.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12962_fsc.xml emd_12962_fsc.xml | 16.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_12962.png emd_12962.png | 86.6 KB | ||
| Filedesc metadata |  emd-12962.cif.gz emd-12962.cif.gz | 5.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12962 http://ftp.pdbj.org/pub/emdb/structures/EMD-12962 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12962 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12962 | HTTPS FTP |
-Validation report
| Summary document |  emd_12962_validation.pdf.gz emd_12962_validation.pdf.gz | 619 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12962_full_validation.pdf.gz emd_12962_full_validation.pdf.gz | 618.6 KB | Display | |
| Data in XML |  emd_12962_validation.xml.gz emd_12962_validation.xml.gz | 15.5 KB | Display | |
| Data in CIF |  emd_12962_validation.cif.gz emd_12962_validation.cif.gz | 21.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12962 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12962 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12962 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12962 | HTTPS FTP |
-Related structure data
| Related structure data |  7oknMC  7okoC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12962.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12962.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of the outer-membrane core complex (inner ring) from a conjugative type IV secretion system | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Outer-membrane core complex (inner ring)
| Entire | Name: Outer-membrane core complex (inner ring) |
|---|---|
| Components |
|
-Supramolecule #1: Outer-membrane core complex (inner ring)
| Supramolecule | Name: Outer-membrane core complex (inner ring) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Salmonella enterica (bacteria) Salmonella enterica (bacteria) |
-Macromolecule #1: TraB
| Macromolecule | Name: TraB / type: protein_or_peptide / ID: 1 / Details: TraB / Number of copies: 17 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella enterica (bacteria) Salmonella enterica (bacteria) |
| Molecular weight | Theoretical: 48.802102 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MANVNKVVRR RQVALLIALV LGIGAGGAGT WMVSEMNLKK APPAKAPKGE PAPDMTGVVN QSFDNKVQRS AIAEAQRLNK ETQTEIKKL RTEMGLVSRD LKGSQDRIRE LEDQNQLLQT QLEAGKNFDS LSAEPLPGAL ASQGKPAPAG NVPPPTSFWP A GGGQAPAA ...String: MANVNKVVRR RQVALLIALV LGIGAGGAGT WMVSEMNLKK APPAKAPKGE PAPDMTGVVN QSFDNKVQRS AIAEAQRLNK ETQTEIKKL RTEMGLVSRD LKGSQDRIRE LEDQNQLLQT QLEAGKNFDS LSAEPLPGAL ASQGKPAPAG NVPPPTSFWP A GGGQAPAA PVMTPIQRPG MMDSQEFSLP DTGPKKPRFP WISSGSFVEA IVVEGADANA SVTGDKNTAP MQLRLTGKVQ MP NDEEFDL TGCFVTLEAW GDVSSERAIV RSRSISCKLG DDDIDQKIAG HVSFMGKNGI KGEVVMRNGQ ILLYAGGAGF LDG IGKGIE KASSTTVGVG ATASMSAADI GQAGLGGGVS SAAKTLSDYY IKRAEQYHPV IPIGAGNEVT LVFQDGFQLE TLEE ARAKA AARKKQNQPS ASSTPAAMPG NTPDMLKQLQ DFRVGDTVDP ATGQVVTQWS HPQFEK |
-Macromolecule #2: Type IV conjugative transfer system lipoprotein TraV
| Macromolecule | Name: Type IV conjugative transfer system lipoprotein TraV / type: protein_or_peptide / ID: 2 / Details: TraV / Number of copies: 17 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella enterica (bacteria) Salmonella enterica (bacteria) |
| Molecular weight | Theoretical: 20.92865 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKKITLLLAG SALLLSGCAG VKSSFDCDAT TSDTCMTMTK ANQLARDKAA KQAGKPAAGG LPSLVNLPAT SAVEVPSASR SAVTPPSGT RTVSTTPPVS AGTSAGVNTN TTTSTLTPRP VAGTPVTTTP SSVAYRPVVS VVTPTPSCQN VRCDNPGTVH P QRSRDQIA ...String: MKKITLLLAG SALLLSGCAG VKSSFDCDAT TSDTCMTMTK ANQLARDKAA KQAGKPAAGG LPSLVNLPAT SAVEVPSASR SAVTPPSGT RTVSTTPPVS AGTSAGVNTN TTTSTLTPRP VAGTPVTTTP SSVAYRPVVS VVTPTPSCQN VRCDNPGTVH P QRSRDQIA TVWIAPWVDS DNAFHQPGRV SFVVSPADWV LPARVN UniProtKB: Type IV conjugative transfer system lipoprotein TraV |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 1.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)