+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | (h-alpha2M)4 transient III state | |||||||||||||||
 Map data Map data | (h-alpha2M)4 transient III state | |||||||||||||||
 Sample Sample |
| |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 9.1 Å | |||||||||||||||
 Authors Authors | Luque D / Goulas T / Mata CP / Mendes SR / Gomis-Ruth FX / Caston JR | |||||||||||||||
| Funding support |  Spain, 4 items Spain, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Cryo-EM structures show the mechanistic basis of pan-peptidase inhibition by human α-macroglobulin. Authors: Daniel Luque / Theodoros Goulas / Carlos P Mata / Soraia R Mendes / F Xavier Gomis-Rüth / José R Castón /    Abstract: Human α2-macroglobulin (hα2M) is a multidomain protein with a plethora of essential functions, including transport of signaling molecules and endopeptidase inhibition in innate immunity. Here, we ...Human α2-macroglobulin (hα2M) is a multidomain protein with a plethora of essential functions, including transport of signaling molecules and endopeptidase inhibition in innate immunity. Here, we dissected the molecular mechanism of the inhibitory function of the ∼720-kDa hα2M tetramer through eight cryo–electron microscopy (cryo-EM) structures of complexes from human plasma. In the native complex, the hα2M subunits are organized in two flexible modules in expanded conformation, which enclose a highly porous cavity in which the proteolytic activity of circulating plasma proteins is tested. Cleavage of bait regions exposed inside the cavity triggers rearrangement to a compact conformation, which closes openings and entraps the prey proteinase. After the expanded-to-compact transition, which occurs independently in the four subunits, the reactive thioester bond triggers covalent linking of the proteinase, and the receptor-binding domain is exposed on the tetramer surface for receptor-mediated clearance from circulation. These results depict the molecular mechanism of a unique suicidal inhibitory trap. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12943.map.gz emd_12943.map.gz | 13 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12943-v30.xml emd-12943-v30.xml emd-12943.xml emd-12943.xml | 13.6 KB 13.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_12943.png emd_12943.png | 186.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12943 http://ftp.pdbj.org/pub/emdb/structures/EMD-12943 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12943 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12943 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12943.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12943.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | (h-alpha2M)4 transient III state | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.047 Å | ||||||||||||||||||||||||||||||||||||
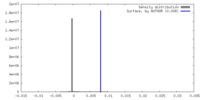
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Human alpha-2-macroglobulin transient III state
| Entire | Name: Human alpha-2-macroglobulin transient III state |
|---|---|
| Components |
|
-Supramolecule #1: Human alpha-2-macroglobulin transient III state
| Supramolecule | Name: Human alpha-2-macroglobulin transient III state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Organ: Blood plasma Homo sapiens (human) / Organ: Blood plasma |
| Molecular weight | Theoretical: 700 KDa |
-Macromolecule #1: Alpha-2-macroglobulin
| Macromolecule | Name: Alpha-2-macroglobulin / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGKNKLLHPS LVLLLLVLLP TDASVSGKPQ YMVLVPSLLH TETTEKGCVL LSYLNETVTV SASLESVRG NRSLFTDLEA ENDVLHCVAF AVPKSSSNEE VMFLTVQVKG PTQEFKKRTT V MVKNEDSL VFVQTDKSIY KPGQTVKFRV VSMDENFHPL NELIPLVYIQ ...String: MGKNKLLHPS LVLLLLVLLP TDASVSGKPQ YMVLVPSLLH TETTEKGCVL LSYLNETVTV SASLESVRG NRSLFTDLEA ENDVLHCVAF AVPKSSSNEE VMFLTVQVKG PTQEFKKRTT V MVKNEDSL VFVQTDKSIY KPGQTVKFRV VSMDENFHPL NELIPLVYIQ DPKGNRIAQW QS FQLEGGL KQFSFPLSSE PFQGSYKVVV QKKSGGRTEH PFTVEEFVLP KFEVQVTVPK IIT ILEEEM NVSVCGLYTY GKPVPGHVTV SICRKYSDAS DCHGEDSQAF CEKFSGQLNS HGCF YQQVK TKVFQLKRKE YEMKLHTEAQ IQEEGTVVEL TGRQSSEITR TITKLSFVKV DSHFR QGIP FFGQVRLVDG KGVPIPNKVI FIRGNEANYY SNATTDEHGL VQFSINTTNV MGTSLT VRV NYKDRSPCYG YQWVSEEHEE AHHTAYLVFS PSKSFVHLEP MSHELPCGHT QTVQAHY IL NGGTLLGLKK LSFYYLIMAK GGIVRTGTHG LLVKQEDMKG HFSISIPVKS DIAPVARL L IYAVLPTGDV IGDSAKYDVE NCLANKVDLS FSPSQSLPAS HAHLRVTAAP QSVCALRAV DQSVLLMKPD AELSASSVYN LLPEKDLTGF PGPLNDQDNE DCINRHNVYI NGITYTPVSS TNEKDMYSF LEDMGLKAFT NSKIRKPKMC PQLQQYEMHG PEGLRVGFYE SDVMGRGHAR L VHVEEPHT ETVRKYFPET WIWDLVVVNS AGVAEVGVTV PDTITEWKAG AFCLSEDAGL GI SSTASLR AFQPFFVELT MPYSVIRGEA FTLKATVLNY LPKCIRVSVQ LEASPAFLAV PVE KEQAPH CICANGRQTV SWAVTPKSLG NVNFTVSAEA LESQELCGTE VPSVPEHGRK DTVI KPLLV EPEGLEKETT FNSLLCPSGG EVSEELSLKL PPNVVEESAR ASVSVLGDIL GSAMQ NTQN LLQMPYGCGE QNMVLFAPNI YVLDYLNETQ QLTPEIKSKA IGYLNTGYQR QLNYKH YDG SYSTFGERYG RNQGNTWLTA FVLKTFAQAR AYIFIDEAHI TQALIWLSQR QKDNGCF RS SGSLLNNAIK GGVEDEVTLS AYITIALLEI PLTVTHPVVR NALFCLESAW KTAQEGDH G SHVYTKALLA YAFALAGNQD KRKEVLKSLN EEAVKKDNSV HWERPQKPKA PVGHFYEPQ APSAEVEMTS YVLLAYLTAQ PAPTSEDLTS ATNIVKWITK QQNAQGGFSS TQDTVVALHA LSKYGAATF TRTGKAAQVT IQSSGTFSSK FQVDNNNRLL LQQVSLPELP GEYSMKVTGE G CVYLQTSL KYNILPEKEE FPFALGVQTL PQTCDEPKAH TSFQISLSVS YTGSRSASNM AI VDVKMVS GFIPLKPTVK MLERSNHVSR TEVSSNHVLI YLDKVSNQTL SLFFTVLQDV PVR DLKPAI VKVYDYYETD EFAIAEYNAP CSKDLGNA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R2/2 / Material: COPPER/RHODIUM |
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA EM CPC |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 12143 / Average electron dose: 39.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.25 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 47775 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)