[English] 日本語
 Yorodumi
Yorodumi- EMDB-12900: Cryo-EM structure of nucleosome core particle composed of the Wid... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12900 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of nucleosome core particle composed of the Widom 601 DNA sequence | |||||||||
 Map data Map data | 3D refinement map from Relion | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | nucleosome / NUCLEAR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationstructural constituent of chromatin / heterochromatin formation / nucleosome / nucleosome assembly / protein heterodimerization activity / DNA binding / nucleoplasm / nucleus Similarity search - Function | |||||||||
| Biological species | ||||||||||
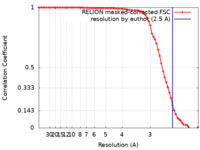
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||
 Authors Authors | Wang H / Cramer P | |||||||||
| Funding support |  Germany, European Union, 2 items Germany, European Union, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: Structures and implications of TBP-nucleosome complexes. Authors: Haibo Wang / Le Xiong / Patrick Cramer /  Abstract: The TATA box-binding protein (TBP) is highly conserved throughout eukaryotes and plays a central role in the assembly of the transcription preinitiation complex (PIC) at gene promoters. TBP binds and ...The TATA box-binding protein (TBP) is highly conserved throughout eukaryotes and plays a central role in the assembly of the transcription preinitiation complex (PIC) at gene promoters. TBP binds and bends DNA, and directs adjacent binding of the transcription factors TFIIA and TFIIB for PIC assembly. Here, we show that yeast TBP can bind to a nucleosome containing the Widom-601 sequence and that TBP-nucleosome binding is stabilized by TFIIA. We determine three cryo-electron microscopy (cryo-EM) structures of TBP-nucleosome complexes, two of them containing also TFIIA. TBP can bind to superhelical location (SHL) -6, which contains a TATA-like sequence, but also to SHL +2, which is GC-rich. Whereas binding to SHL -6 can occur in the absence of TFIIA, binding to SHL +2 is only observed in the presence of TFIIA and goes along with detachment of upstream terminal DNA from the histone octamer. TBP-nucleosome complexes are sterically incompatible with PIC assembly, explaining why a promoter nucleosome generally impairs transcription and must be moved before initiation can occur. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12900.map.gz emd_12900.map.gz | 23.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12900-v30.xml emd-12900-v30.xml emd-12900.xml emd-12900.xml | 18.5 KB 18.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12900_fsc.xml emd_12900_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_12900.png emd_12900.png | 249.7 KB | ||
| Filedesc metadata |  emd-12900.cif.gz emd-12900.cif.gz | 6.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12900 http://ftp.pdbj.org/pub/emdb/structures/EMD-12900 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12900 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12900 | HTTPS FTP |
-Related structure data
| Related structure data |  7ohcMC  7oh9C  7ohaC  7ohbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12900.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12900.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D refinement map from Relion | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Nucleosome core particle
| Entire | Name: Nucleosome core particle |
|---|---|
| Components |
|
-Supramolecule #1: Nucleosome core particle
| Supramolecule | Name: Nucleosome core particle / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Molecular weight | Theoretical: 200 KDa |
-Macromolecule #1: Histone H3.2
| Macromolecule | Name: Histone H3.2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 15.30393 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ARTKQTARKS TGGKAPRKQL ATKAARKSAP ATGGVKKPHR YRPGTVALRE IRRYQKSTEL LIRKLPFQRL VREIAQDFKT DLRFQSSAV MALQEASEAY LVALFEDTNL CAIHAKRVTI MPKDIQLARR IRGERA UniProtKB: Histone H3.2 |
-Macromolecule #2: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 11.263231 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SGRGKGGKGL GKGGAKRHRK VLRDNIQGIT KPAIRRLARR GGVKRISGLI YEETRGVLKV FLENVIRDAV TYTEHAKRKT VTAMDVVYA LKRQGRTLYG FGG UniProtKB: Histone H4 |
-Macromolecule #3: Histone H2A
| Macromolecule | Name: Histone H2A / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 13.978241 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SGRGKQGGKT RAKAKTRSSR AGLQFPVGRV HRLLRKGNYA ERVGAGAPVY LAAVLEYLTA EILELAGNAA RDNKKTRIIP RHLQLAVRN DEELNKLLGR VTIAQGGVLP NIQSVLLPKK TESSKSAKSK UniProtKB: Histone H2A |
-Macromolecule #4: Histone H2B 1.1
| Macromolecule | Name: Histone H2B 1.1 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 13.524752 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AKSAPAPKKG SKKAVTKTQK KDGKKRRKTR KESYAIYVYK VLKQVHPDTG ISSKAMSIMN SFVNDVFERI AGEASRLAHY NKRSTITSR EIQTAVRLLL PGELAKHAVS EGTKAVTKYT SAK UniProtKB: Histone H2B 1.1 |
-Macromolecule #5: DNA (145-MER)
| Macromolecule | Name: DNA (145-MER) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 44.520383 KDa |
| Sequence | String: (DA)(DT)(DC)(DA)(DG)(DA)(DA)(DT)(DC)(DC) (DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA)(DG) (DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA)(DA) (DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA)(DG) (DA) (DC)(DA)(DG)(DC)(DT)(DC) ...String: (DA)(DT)(DC)(DA)(DG)(DA)(DA)(DT)(DC)(DC) (DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA)(DG) (DG)(DC)(DC)(DG)(DC)(DT)(DC)(DA)(DA) (DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA)(DG) (DA) (DC)(DA)(DG)(DC)(DT)(DC)(DT)(DA) (DG)(DC)(DA)(DC)(DC)(DG)(DC)(DT)(DT)(DA) (DA)(DA) (DC)(DG)(DC)(DA)(DC)(DG)(DT) (DA)(DC)(DG)(DC)(DG)(DC)(DT)(DG)(DT)(DC) (DC)(DC)(DC) (DC)(DG)(DC)(DG)(DT)(DT) (DT)(DT)(DA)(DA)(DC)(DC)(DG)(DC)(DC)(DA) (DA)(DG)(DG)(DG) (DG)(DA)(DT)(DT)(DA) (DC)(DT)(DC)(DC)(DC)(DT)(DA)(DG)(DT)(DC) (DT)(DC)(DC)(DA)(DG) (DG)(DC)(DA)(DC) (DG)(DT)(DG)(DT)(DC)(DA)(DG)(DA)(DT)(DA) (DT)(DA)(DT)(DA)(DC)(DA) (DT)(DC)(DG) (DA)(DT) |
-Macromolecule #6: DNA (145-MER)
| Macromolecule | Name: DNA (145-MER) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 44.99166 KDa |
| Sequence | String: (DA)(DT)(DC)(DG)(DA)(DT)(DG)(DT)(DA)(DT) (DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC)(DA) (DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG)(DG) (DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG)(DA) (DG) (DT)(DA)(DA)(DT)(DC)(DC) ...String: (DA)(DT)(DC)(DG)(DA)(DT)(DG)(DT)(DA)(DT) (DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC)(DA) (DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG)(DG) (DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG)(DA) (DG) (DT)(DA)(DA)(DT)(DC)(DC)(DC)(DC) (DT)(DT)(DG)(DG)(DC)(DG)(DG)(DT)(DT)(DA) (DA)(DA) (DA)(DC)(DG)(DC)(DG)(DG)(DG) (DG)(DG)(DA)(DC)(DA)(DG)(DC)(DG)(DC)(DG) (DT)(DA)(DC) (DG)(DT)(DG)(DC)(DG)(DT) (DT)(DT)(DA)(DA)(DG)(DC)(DG)(DG)(DT)(DG) (DC)(DT)(DA)(DG) (DA)(DG)(DC)(DT)(DG) (DT)(DC)(DT)(DA)(DC)(DG)(DA)(DC)(DC)(DA) (DA)(DT)(DT)(DG)(DA) (DG)(DC)(DG)(DG) (DC)(DC)(DT)(DC)(DG)(DG)(DC)(DA)(DC)(DC) (DG)(DG)(DG)(DA)(DT)(DT) (DC)(DT)(DG) (DA)(DT) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.12 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-7ohc: |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)