[English] 日本語
 Yorodumi
Yorodumi- EMDB-12823: Hepatitis B core protein capsid, mutant F97L with bound MHRSLLGRMKGA -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12823 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Hepatitis B core protein capsid, mutant F97L with bound MHRSLLGRMKGA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationmicrotubule-dependent intracellular transport of viral material towards nucleus / T=4 icosahedral viral capsid / viral penetration into host nucleus / host cell / host cell cytoplasm / symbiont entry into host cell / structural molecule activity / DNA binding / RNA binding Similarity search - Function | |||||||||
| Biological species |   Hepatitis B virus / synthetic construct (others) Hepatitis B virus / synthetic construct (others) | |||||||||
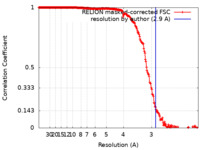
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Bottcher B / Makbul C | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Microorganisms / Year: 2021 Journal: Microorganisms / Year: 2021Title: Conformational Plasticity of Hepatitis B Core Protein Spikes Promotes Peptide Binding Independent of the Secretion Phenotype. Authors: Cihan Makbul / Vladimir Khayenko / Hans Michael Maric / Bettina Böttcher /  Abstract: Hepatitis B virus is a major human pathogen, which forms enveloped virus particles. During viral maturation, membrane-bound hepatitis B surface proteins package hepatitis B core protein capsids. This ...Hepatitis B virus is a major human pathogen, which forms enveloped virus particles. During viral maturation, membrane-bound hepatitis B surface proteins package hepatitis B core protein capsids. This process is intercepted by certain peptides with an "LLGRMKG" motif that binds to the capsids at the tips of dimeric spikes. With microcalorimetry, electron cryo microscopy and peptide microarray-based screens, we have characterized the structural and thermodynamic properties of peptide binding to hepatitis B core protein capsids with different secretion phenotypes. The peptide "GSLLGRMKGA" binds weakly to hepatitis B core protein capsids and mutant capsids with a premature (F97L) or low-secretion phenotype (L60V and P5T). With electron cryo microscopy, we provide novel structures for L60V and P5T and demonstrate that binding occurs at the tips of the spikes at the dimer interface, splaying the helices apart independent of the secretion phenotype. Peptide array screening identifies "SLLGRM" as the core binding motif. This shortened motif binds only to one of the two spikes in the asymmetric unit of the capsid and induces a much smaller conformational change. Altogether, these comprehensive studies suggest that the tips of the spikes act as an autonomous binding platform that is unaffected by mutations that affect secretion phenotypes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12823.map.gz emd_12823.map.gz | 227 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12823-v30.xml emd-12823-v30.xml emd-12823.xml emd-12823.xml | 16.4 KB 16.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12823_fsc.xml emd_12823_fsc.xml | 21.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_12823.png emd_12823.png | 211.4 KB | ||
| Others |  emd_12823_half_map_1.map.gz emd_12823_half_map_1.map.gz emd_12823_half_map_2.map.gz emd_12823_half_map_2.map.gz | 668.3 MB 668.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12823 http://ftp.pdbj.org/pub/emdb/structures/EMD-12823 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12823 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12823 | HTTPS FTP |
-Validation report
| Summary document |  emd_12823_validation.pdf.gz emd_12823_validation.pdf.gz | 362.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12823_full_validation.pdf.gz emd_12823_full_validation.pdf.gz | 361.4 KB | Display | |
| Data in XML |  emd_12823_validation.xml.gz emd_12823_validation.xml.gz | 25.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12823 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12823 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12823 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12823 | HTTPS FTP |
-Related structure data
| Related structure data |  7oewMC  7ocoC  7ocwC  7od4C  7od6C  7od7C  7od8C  7oenC  7oevC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12823.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12823.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0635 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
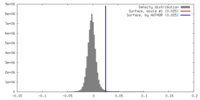
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_12823_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
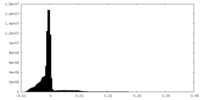
| Density Histograms |
-Half map: #2
| File | emd_12823_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
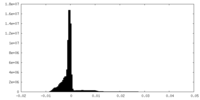
| Density Histograms |
- Sample components
Sample components
-Entire : Hepatitis B virus
| Entire | Name:   Hepatitis B virus Hepatitis B virus |
|---|---|
| Components |
|
-Supramolecule #1: Hepatitis B virus
| Supramolecule | Name: Hepatitis B virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 10407 / Sci species name: Hepatitis B virus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host system | Organism:  |
| Molecular weight | Theoretical: 4.8 MDa |
| Virus shell | Shell ID: 1 / Name: capsid / Diameter: 360.0 Å / T number (triangulation number): 4 |
-Macromolecule #1: Hepatitis B core protein
| Macromolecule | Name: Hepatitis B core protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Hepatitis B virus Hepatitis B virus |
| Recombinant expression | Organism:  |
| Sequence | String: MDIDPYKEFG ATVELLSFLP SDFFPSVRDL LDTASALYRE ALESPEHCSP HHTALRQAIL CWGELMTLA TWVGVNLEDP ASRDLVVSYV NTNMGLKLRQ LLWFHISCLT FGRETVIEYL V SFGVWIRT PPAYRPPNAP ILSTLPETTV VRRRGRSPRR RTPSPRRRRS QSPRRRRSQS RE SQC |
-Macromolecule #2: Peptide MHRSLLGRMKGA
| Macromolecule | Name: Peptide MHRSLLGRMKGA / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Sequence | String: MHRSLLGRMK GA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: PLASMA CLEANING |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Number grids imaged: 1 / Number real images: 1110 / Average exposure time: 5.0 sec. / Average electron dose: 8.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 95 |
|---|---|
| Output model |  PDB-7oew: |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)