[English] 日本語
 Yorodumi
Yorodumi- EMDB-12652: Respiratory complex I from Escherichia coli - focused refinement ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12652 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Respiratory complex I from Escherichia coli - focused refinement of membrane arm | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | NADH:ubiquinone reductase (H+-translocating) / oxidative phosphorylation / ELECTRON TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationTranslocases; Catalysing the translocation of protons; Linked to oxidoreductase reactions / NADH dehydrogenase (quinone) (non-electrogenic) activity / NADH dehydrogenase complex / oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor / ubiquinone binding / electron transport coupled proton transport / respiratory chain complex I / membrane => GO:0016020 / NADH dehydrogenase (ubiquinone) activity / quinone binding ...Translocases; Catalysing the translocation of protons; Linked to oxidoreductase reactions / NADH dehydrogenase (quinone) (non-electrogenic) activity / NADH dehydrogenase complex / oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor / ubiquinone binding / electron transport coupled proton transport / respiratory chain complex I / membrane => GO:0016020 / NADH dehydrogenase (ubiquinone) activity / quinone binding / ATP synthesis coupled electron transport / proton transmembrane transport / aerobic respiration / respiratory electron transport chain / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
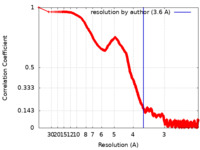
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Kolata P / Efremov RG | |||||||||
| Funding support |  Belgium, 1 items Belgium, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2021 Journal: Elife / Year: 2021Title: Structure of respiratory complex I reconstituted into lipid nanodiscs reveals an uncoupled conformation. Authors: Piotr Kolata / Rouslan G Efremov /  Abstract: Respiratory complex I is a multi-subunit membrane protein complex that reversibly couples NADH oxidation and ubiquinone reduction with proton translocation against transmembrane potential. Complex I ...Respiratory complex I is a multi-subunit membrane protein complex that reversibly couples NADH oxidation and ubiquinone reduction with proton translocation against transmembrane potential. Complex I from is among the best functionally characterized complexes, but its structure remains unknown, hindering further studies to understand the enzyme coupling mechanism. Here, we describe the single particle cryo-electron microscopy (cryo-EM) structure of the entire catalytically active complex I reconstituted into lipid nanodiscs. The structure of this mesophilic bacterial complex I displays highly dynamic connection between the peripheral and membrane domains. The peripheral domain assembly is stabilized by unique terminal extensions and an insertion loop. The membrane domain structure reveals novel dynamic features. Unusual conformation of the conserved interface between the peripheral and membrane domains suggests an uncoupled conformation of the complex. Considering constraints imposed by the structural data, we suggest a new simple hypothetical coupling mechanism for the molecular machine. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12652.map.gz emd_12652.map.gz | 4.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12652-v30.xml emd-12652-v30.xml emd-12652.xml emd-12652.xml | 27.2 KB 27.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12652_fsc.xml emd_12652_fsc.xml | 267.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_12652.png emd_12652.png | 128.4 KB | ||
| Filedesc metadata |  emd-12652.cif.gz emd-12652.cif.gz | 7.7 KB | ||
| Others |  emd_12652_half_map_1.map.gz emd_12652_half_map_1.map.gz emd_12652_half_map_2.map.gz emd_12652_half_map_2.map.gz | 98.6 MB 98.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12652 http://ftp.pdbj.org/pub/emdb/structures/EMD-12652 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12652 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12652 | HTTPS FTP |
-Related structure data
| Related structure data |  7nyhMC  7nyrC  7nyuC  7nyvC  7nz1C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12652.map.gz / Format: CCP4 / Size: 5.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12652.map.gz / Format: CCP4 / Size: 5.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1568 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #2
| File | emd_12652_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
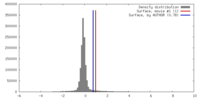
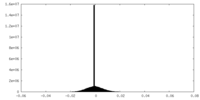
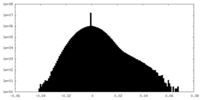
| Density Histograms |
-Half map: #1
| File | emd_12652_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
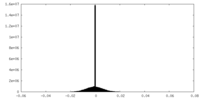
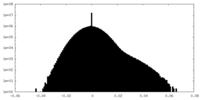
| Density Histograms |
- Sample components
Sample components
-Entire : Respiratory complex I from Escherichia coli - focused refinement ...
| Entire | Name: Respiratory complex I from Escherichia coli - focused refinement of membrane arm |
|---|---|
| Components |
|
-Supramolecule #1: Respiratory complex I from Escherichia coli - focused refinement ...
| Supramolecule | Name: Respiratory complex I from Escherichia coli - focused refinement of membrane arm type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 260 KDa |
-Macromolecule #1: NADH-quinone oxidoreductase subunit A
| Macromolecule | Name: NADH-quinone oxidoreductase subunit A / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Translocases; Catalysing the translocation of protons; Linked to oxidoreductase reactions |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.474283 KDa |
| Sequence | String: MSMSTSTEVI AHHWAFAIFL IVAIGLCCLM LVGGWFLGGR ARARSKNVPF ESGIDSVGSA RLRLSAKFYL VAMFFVIFDV EALYLFAWS TSIRESGWVG FVEAAIFIFV LLAGLVYLVR IGALDWTPAR SRRERMNPET NSIANRQR UniProtKB: NADH-quinone oxidoreductase subunit A |
-Macromolecule #2: NADH-quinone oxidoreductase subunit H
| Macromolecule | Name: NADH-quinone oxidoreductase subunit H / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number: Translocases; Catalysing the translocation of protons; Linked to oxidoreductase reactions |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 36.240922 KDa |
| Sequence | String: MSWISPELIE ILLTILKAVV ILLVVVTCGA FMSFGERRLL GLFQNRYGPN RVGWGGSLQL VADMIKMFFK EDWIPKFSDR VIFTLAPMI AFTSLLLAFA IVPVSPGWVV ADLNIGILFF LMMAGLAVYA VLFAGWSSNN KYSLLGAMRA SAQTLSYEVF L GLSLMGVV ...String: MSWISPELIE ILLTILKAVV ILLVVVTCGA FMSFGERRLL GLFQNRYGPN RVGWGGSLQL VADMIKMFFK EDWIPKFSDR VIFTLAPMI AFTSLLLAFA IVPVSPGWVV ADLNIGILFF LMMAGLAVYA VLFAGWSSNN KYSLLGAMRA SAQTLSYEVF L GLSLMGVV AQAGSFNMTD IVNSQAHVWN VIPQFFGFIT FAIAGVAVCH RHPFDQPEAE QELADGYHIE YSGMKFGLFF VG EYIGIVT ISALMVTLFF GGWQGPLLPP FIWFALKTAF FMMMFILIRA SLPRPRYDQV MSFGWKICLP LTLINLLVTA AVI LWQAQ UniProtKB: NADH-quinone oxidoreductase subunit H |
-Macromolecule #3: NADH-quinone oxidoreductase subunit J
| Macromolecule | Name: NADH-quinone oxidoreductase subunit J / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO EC number: Translocases; Catalysing the translocation of protons; Linked to oxidoreductase reactions |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 19.889551 KDa |
| Sequence | String: MEFAFYICGL IAILATLRVI THTNPVHALL YLIISLLAIS GVFFSLGAYF AGALEIIVYA GAIMVLFVFV VMMLNLGGSE IEQERQWLK PQVWIGPAIL SAIMLVVIVY AILGVNDQGI DGTPISAKAV GITLFGPYVL AVELASMLLL AGLVVAFHVG R EERAGEVL SNRKDDSAKR KTEEHA UniProtKB: NADH-quinone oxidoreductase subunit J |
-Macromolecule #4: NADH-quinone oxidoreductase subunit K
| Macromolecule | Name: NADH-quinone oxidoreductase subunit K / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO / EC number: NADH:ubiquinone reductase (H+-translocating) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 10.852961 KDa |
| Sequence | String: MIPLQHGLIL AAILFVLGLT GLVIRRNLLF MLIGLEIMIN ASALAFVVAG SYWGQTDGQV MYILAISLAA AEASIGLALL LQLHRRRQN LNIDSVSEMR G UniProtKB: NADH-quinone oxidoreductase subunit K |
-Macromolecule #5: NADH-quinone oxidoreductase subunit L
| Macromolecule | Name: NADH-quinone oxidoreductase subunit L / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO / EC number: NADH dehydrogenase (quinone) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 66.513633 KDa |
| Sequence | String: MNMLALTIIL PLIGFVLLAF SRGRWSENVS AIVGVGSVGL AALVTAFIGV DFFANGEQTY SQPLWTWMSV GDFNIGFNLV LDGLSLTML SVVTGVGFLI HMYASWYMRG EEGYSRFFAY TNLFIASMVV LVLADNLLLM YLGWEGVGLC SYLLIGFYYT D PKNGAAAM ...String: MNMLALTIIL PLIGFVLLAF SRGRWSENVS AIVGVGSVGL AALVTAFIGV DFFANGEQTY SQPLWTWMSV GDFNIGFNLV LDGLSLTML SVVTGVGFLI HMYASWYMRG EEGYSRFFAY TNLFIASMVV LVLADNLLLM YLGWEGVGLC SYLLIGFYYT D PKNGAAAM KAFVVTRVGD VFLAFALFIL YNELGTLNFR EMVELAPAHF ADGNNMLMWA TLMLLGGAVG KSAQLPLQTW LA DAMAGPT PVSALIHAAT MVTAGVYLIA RTHGLFLMTP EVLHLVGIVG AVTLLLAGFA ALVQTDIKRV LAYSTMSQIG YMF LALGVQ AWDAAIFHLM THAFFKALLF LASGSVILAC HHEQNIFKMG GLRKSIPLVY LCFLVGGAAL SALPLVTAGF FSKD EILAG AMANGHINLM VAGLVGAFMT SLYTFRMIFI VFHGKEQIHA HAVKGVTHSL PLIVLLILST FVGALIVPPL QGVLP QTTE LAHGSMLTLE ITSGVVAVVG ILLAAWLWLG KRTLVTSIAN SAPGRLLSTW WYNAWGFDWL YDKVFVKPFL GIAWLL KRD PLNSMMNIPA VLSRFAGKGL LLSENGYLRW YVASMSIGAV VVLALLMVLR UniProtKB: NADH-quinone oxidoreductase subunit L |
-Macromolecule #6: NADH-quinone oxidoreductase subunit M
| Macromolecule | Name: NADH-quinone oxidoreductase subunit M / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO EC number: Translocases; Catalysing the translocation of protons; Linked to oxidoreductase reactions |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 56.56009 KDa |
| Sequence | String: MLLPWLILIP FIGGFLCWQT ERFGVKVPRW IALITMGLTL ALSLQLWLQG GYSLTQSAGI PQWQSEFDMP WIPRFGISIH LAIDGLSLL MVVLTGLLGV LAVLCSWKEI EKYQGFFHLN LMWILGGVIG VFLAIDMFLF FFFWEMMLVP MYFLIALWGH K ASDGKTRI ...String: MLLPWLILIP FIGGFLCWQT ERFGVKVPRW IALITMGLTL ALSLQLWLQG GYSLTQSAGI PQWQSEFDMP WIPRFGISIH LAIDGLSLL MVVLTGLLGV LAVLCSWKEI EKYQGFFHLN LMWILGGVIG VFLAIDMFLF FFFWEMMLVP MYFLIALWGH K ASDGKTRI TAATKFFIYT QASGLVMLIA ILALVFVHYN ATGVWTFNYE ELLNTPMSSG VEYLLMLGFF IAFAVKMPVV PL HGWLPDA HSQAPTAGSV DLAGILLKTA AYGLLRFSLP LFPNASAEFA PIAMWLGVIG IFYGAWMAFA QTDIKRLIAY TSV SHMGFV LIAIYTGSQL AYQGAVIQMI AHGLSAAGLF ILCGQLYERI HTRDMRMMGG LWSKMKWLPA LSLFFAVATL GMPG TGNFV GEFMILFGSF QVVPVITVIS TFGLVFASVY SLAMLHRAYF GKAKSQIASQ ELPGMSLREL FMILLLVVLL VLLGF YPQP ILDTSHSAIG NIQQWFVNSV TTTRP UniProtKB: NADH-quinone oxidoreductase subunit M |
-Macromolecule #7: NADH-quinone oxidoreductase subunit N
| Macromolecule | Name: NADH-quinone oxidoreductase subunit N / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO EC number: Translocases; Catalysing the translocation of protons; Linked to oxidoreductase reactions |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 52.072672 KDa |
| Sequence | String: MTITPQNLIA LLPLLIVGLT VVVVMLSIAW RRNHFLNATL SVIGLNAALV SLWFVGQAGA MDVTPLMRVD GFAMLYTGLV LLASLATCT FAYPWLEGYN DNKDEFYLLV LIAALGGILL ANANHLASLF LGIELISLPL FGLVGYAFRQ KRSLEASIKY T ILSAAASS ...String: MTITPQNLIA LLPLLIVGLT VVVVMLSIAW RRNHFLNATL SVIGLNAALV SLWFVGQAGA MDVTPLMRVD GFAMLYTGLV LLASLATCT FAYPWLEGYN DNKDEFYLLV LIAALGGILL ANANHLASLF LGIELISLPL FGLVGYAFRQ KRSLEASIKY T ILSAAASS FLLFGMALVY AQSGDLSFVA LGKNLGDGML NEPLLLAGFG LMIVGLGFKL SLVPFHLWTP DVYQGAPAPV ST FLATASK IAIFGVVMRL FLYAPVGDSE AIRVVLAIIA FASIIFGNLM ALSQTNIKRL LGYSSISHLG YLLVALIALQ TGE MSMEAV GVYLAGYLFS SLGAFGVVSL MSSPYRGPDA DSLFSYRGLF WHRPILAAVM TVMMLSLAGI PMTLGFIGKF YVLA VGVQA HLWWLVGAVV VGSAIGLYYY LRVAVSLYLH APEQPGRDAP SNWQYSAGGI VVLISALLVL VLGVWPQPLI SIVRL AMPL M UniProtKB: NADH-quinone oxidoreductase subunit N |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.8 Component:
Details: The buffer was used for gel filtration of protein reconstituted in lipid nanodiscs | ||||||||||||
| Grid | Model: Quantifoil R0.6/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 12 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 240 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.028 kPa | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 97 % / Chamber temperature: 296 K / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 9122 / Average exposure time: 3.0 sec. / Average electron dose: 64.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.55 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: JEOL CRYOSPECPORTER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 75 / Target criteria: Correlation coefficient | ||||||
| Output model |  PDB-7nyh: |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)