+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12595 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rhinovirus-14 ICAM-1 empty particle at pH 6.2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | rhinovirus 14 / RV14 / HRV14 / acidification / pH 6.2 / genome release / emptu / ICAM-1 / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationlysis of host organelle involved in viral entry into host cell / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity ...lysis of host organelle involved in viral entry into host cell / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / DNA replication / RNA helicase activity / endocytosis involved in viral entry into host cell / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / ATP hydrolysis activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  Human rhinovirus 14 / Human rhinovirus 14 /  Rhinovirus B14 Rhinovirus B14 | |||||||||
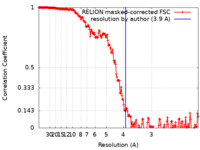
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Hrebik D / Plevka P | |||||||||
| Funding support |  Czech Republic, 1 items Czech Republic, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: ICAM-1 induced rearrangements of capsid and genome prime rhinovirus 14 for activation and uncoating. Authors: Dominik Hrebík / Tibor Füzik / Mária Gondová / Lenka Šmerdová / Athanassios Adamopoulos / Ondrej Šedo / Zbyněk Zdráhal / Pavel Plevka /  Abstract: Most rhinoviruses, which are the leading cause of the common cold, utilize intercellular adhesion molecule-1 (ICAM-1) as a receptor to infect cells. To release their genomes, rhinoviruses convert to ...Most rhinoviruses, which are the leading cause of the common cold, utilize intercellular adhesion molecule-1 (ICAM-1) as a receptor to infect cells. To release their genomes, rhinoviruses convert to activated particles that contain pores in the capsid, lack minor capsid protein VP4, and have an altered genome organization. The binding of rhinoviruses to ICAM-1 promotes virus activation; however, the molecular details of the process remain unknown. Here, we present the structures of virion of rhinovirus 14 and its complex with ICAM-1 determined to resolutions of 2.6 and 2.4 Å, respectively. The cryo-electron microscopy reconstruction of rhinovirus 14 virions contains the resolved density of octanucleotide segments from the RNA genome that interact with VP2 subunits. We show that the binding of ICAM-1 to rhinovirus 14 is required to prime the virus for activation and genome release at acidic pH. Formation of the rhinovirus 14-ICAM-1 complex induces conformational changes to the rhinovirus 14 capsid, including translocation of the C termini of VP4 subunits, which become poised for release through pores that open in the capsids of activated particles. VP4 subunits with altered conformation block the RNA-VP2 interactions and expose patches of positively charged residues. The conformational changes to the capsid induce the redistribution of the virus genome by altering the capsid-RNA interactions. The restructuring of the rhinovirus 14 capsid and genome prepares the virions for conversion to activated particles. The high-resolution structure of rhinovirus 14 in complex with ICAM-1 explains how the binding of uncoating receptors enables enterovirus genome release. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12595.map.gz emd_12595.map.gz | 403.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12595-v30.xml emd-12595-v30.xml emd-12595.xml emd-12595.xml | 22.2 KB 22.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12595_fsc.xml emd_12595_fsc.xml | 18.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_12595.png emd_12595.png | 354.7 KB | ||
| Masks |  emd_12595_msk_1.map emd_12595_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-12595.cif.gz emd-12595.cif.gz | 6.6 KB | ||
| Others |  emd_12595_half_map_1.map.gz emd_12595_half_map_1.map.gz emd_12595_half_map_2.map.gz emd_12595_half_map_2.map.gz | 409.5 MB 409 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12595 http://ftp.pdbj.org/pub/emdb/structures/EMD-12595 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12595 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12595 | HTTPS FTP |
-Validation report
| Summary document |  emd_12595_validation.pdf.gz emd_12595_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12595_full_validation.pdf.gz emd_12595_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_12595_validation.xml.gz emd_12595_validation.xml.gz | 26.7 KB | Display | |
| Data in CIF |  emd_12595_validation.cif.gz emd_12595_validation.cif.gz | 35.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12595 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12595 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12595 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12595 | HTTPS FTP |
-Related structure data
| Related structure data |  7numMC  7bg6C  7bg7C  7nulC  7nunC  7nuoC  7nuqC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12595.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12595.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.063 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
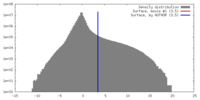
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12595_msk_1.map emd_12595_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_12595_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_12595_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Rhinovirus B14
| Entire | Name:  Rhinovirus B14 Rhinovirus B14 |
|---|---|
| Components |
|
-Supramolecule #1: Rhinovirus B14
| Supramolecule | Name: Rhinovirus B14 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 12131 / Sci species name: Rhinovirus B14 / Sci species strain: 1059 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7 MDa |
| Virus shell | Shell ID: 1 / Name: Rhinovirus B14 / Diameter: 295.0 Å / T number (triangulation number): 3 |
-Macromolecule #1: Genome polyprotein
| Macromolecule | Name: Genome polyprotein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: picornain 2A |
|---|---|
| Source (natural) | Organism:  Human rhinovirus 14 Human rhinovirus 14 |
| Molecular weight | Theoretical: 32.975004 KDa |
| Sequence | String: ALTEGLGDEL EEVIVEKTKQ TVASISSGPK HTQKVPILTA NETGATMPVL PSDSIETRTT YMHFNGSETD VECFLGRAAC VHVTEIQNK DATGIDNHRE AKLFNDWKIN LSSLVQLRKK LELFTYVRFD SEYTILATAS QPDSANYSSN LVVQAMYVPP G APNPKEWD ...String: ALTEGLGDEL EEVIVEKTKQ TVASISSGPK HTQKVPILTA NETGATMPVL PSDSIETRTT YMHFNGSETD VECFLGRAAC VHVTEIQNK DATGIDNHRE AKLFNDWKIN LSSLVQLRKK LELFTYVRFD SEYTILATAS QPDSANYSSN LVVQAMYVPP G APNPKEWD DYTWQSASNP SVFFKVGDTS RFSVPYVGLA SAYNCFYDGY SHDDAETQYG ITVLNHMGSM AFRIVNEHDE HK TLVKIRV YHRAKHVEAW IPRAPRALPY TSIGRTNYPK NTEPVIKKRK GDIKSY UniProtKB: Genome polyprotein |
-Macromolecule #2: P1
| Macromolecule | Name: P1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human rhinovirus 14 Human rhinovirus 14 |
| Molecular weight | Theoretical: 25.822299 KDa |
| Sequence | String: GLPTTTLPGS GQFLTTDDRQ SPSALPNYEP TPRIHIPGKV HNLLEIIQVD TLIPMNNTHT KDEVNSYLIP LNANRQNEQV FGTNLFIGD GVFKTTLLGE IVQYYTHWSG SLRFSLMYTG PALSSAKLIL AYTPPGARGP QDRREAMLGT HVVWDIGLQS T IVMTIPWT ...String: GLPTTTLPGS GQFLTTDDRQ SPSALPNYEP TPRIHIPGKV HNLLEIIQVD TLIPMNNTHT KDEVNSYLIP LNANRQNEQV FGTNLFIGD GVFKTTLLGE IVQYYTHWSG SLRFSLMYTG PALSSAKLIL AYTPPGARGP QDRREAMLGT HVVWDIGLQS T IVMTIPWT SGVQFRYTDP DTYTSAGFLS CWYQTSLILP PETTGQVYLL SFISACPDFK LRLMKDTQTI SQTV UniProtKB: Genome polyprotein |
-Macromolecule #3: Genome polyprotein
| Macromolecule | Name: Genome polyprotein / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: picornain 2A |
|---|---|
| Source (natural) | Organism:  Human rhinovirus 14 Human rhinovirus 14 |
| Molecular weight | Theoretical: 28.501361 KDa |
| Sequence | String: SPNVEACGYS DRVQQITLGN STITTQEAAN AVVCYAEWPE YLPDVDASDV NKTSKPDTSV CRFYTLDSKT WTTGSKGWCW KLPDALKDM GVFGQNMFFH SLGRSGYTVH VQCNATKFHS GCLLVVVIPE HQLASHEGGN VSVKYTFTHP GERGIDLSSA N EVGGPVKD ...String: SPNVEACGYS DRVQQITLGN STITTQEAAN AVVCYAEWPE YLPDVDASDV NKTSKPDTSV CRFYTLDSKT WTTGSKGWCW KLPDALKDM GVFGQNMFFH SLGRSGYTVH VQCNATKFHS GCLLVVVIPE HQLASHEGGN VSVKYTFTHP GERGIDLSSA N EVGGPVKD VIYNMNGTLL GNLLIFPHQF INLRTNNTAT IVIPYINSVP IDSMTRHNNV SLMVIPIAPL TVPTGATPSL PI TVTIAPM CTEFSGIRSK SIVPQ UniProtKB: Genome polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 6.2 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: OTHER | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 279.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 6548 / Average exposure time: 1.0 sec. / Average electron dose: 77.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 3.95 µm / Calibrated defocus min: 0.25 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)