[English] 日本語
 Yorodumi
Yorodumi- EMDB-12483: Cryo-EM structure of the folate-specific ECF transporter complex ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12483 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the folate-specific ECF transporter complex in DDM micelles | |||||||||||||||
 Map data Map data | Inward-facing apo conformation of the folate-specific ECF transporter complex in DDM micelles at 3.4 A resolution sharpened at -62 A^2. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | ABC Transporter / Type III ABC Transporter / ECF transporter complex / Folate transporter / Membrane protein / TRANSPORT PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationTranslocases / ATPase-coupled transmembrane transporter activity / transmembrane transporter activity / ATP-binding cassette (ABC) transporter complex / ATP hydrolysis activity / ATP binding / membrane / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  Lactobacillus delbrueckii subsp. bulgaricus (strain ATCC 11842 / DSM 20081 / JCM 1002 / NBRC 13953 / NCIMB 11778) (bacteria) Lactobacillus delbrueckii subsp. bulgaricus (strain ATCC 11842 / DSM 20081 / JCM 1002 / NBRC 13953 / NCIMB 11778) (bacteria) | |||||||||||||||
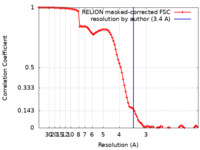
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||||||||
 Authors Authors | Thangaratnarajah C / Rheinberger J | |||||||||||||||
| Funding support |  Netherlands, 4 items Netherlands, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: Insights into the bilayer-mediated toppling mechanism of a folate-specific ECF transporter by cryo-EM. Authors: Chancievan Thangaratnarajah / Jan Rheinberger / Cristina Paulino / Dirk J Slotboom /  Abstract: Energy-coupling factor (ECF)-type transporters are small, asymmetric membrane protein complexes (∼115 kDa) that consist of a membrane-embedded, substrate-binding protein (S component) and a ...Energy-coupling factor (ECF)-type transporters are small, asymmetric membrane protein complexes (∼115 kDa) that consist of a membrane-embedded, substrate-binding protein (S component) and a tripartite ATP-hydrolyzing module (ECF module). They import micronutrients into bacterial cells and have been proposed to use a highly unusual transport mechanism, in which the substrate is dragged across the membrane by a toppling motion of the S component. However, it remains unclear how the lipid bilayer could accommodate such a movement. Here, we used cryogenic electron microscopy at 200 kV to determine structures of a folate-specific ECF transporter in lipid nanodiscs and detergent micelles at 2.7- and 3.4-Å resolution, respectively. The structures reveal an irregularly shaped bilayer environment around the membrane-embedded complex and suggest that toppling of the S component is facilitated by protein-induced membrane deformations. In this way, structural remodeling of the lipid bilayer environment is exploited to guide the transport process. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12483.map.gz emd_12483.map.gz | 59.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12483-v30.xml emd-12483-v30.xml emd-12483.xml emd-12483.xml | 27 KB 27 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12483_fsc.xml emd_12483_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_12483.png emd_12483.png | 158.6 KB | ||
| Masks |  emd_12483_msk_1.map emd_12483_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-12483.cif.gz emd-12483.cif.gz | 7.3 KB | ||
| Others |  emd_12483_additional_1.map.gz emd_12483_additional_1.map.gz emd_12483_half_map_1.map.gz emd_12483_half_map_1.map.gz emd_12483_half_map_2.map.gz emd_12483_half_map_2.map.gz | 49.3 MB 49.6 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12483 http://ftp.pdbj.org/pub/emdb/structures/EMD-12483 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12483 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12483 | HTTPS FTP |
-Related structure data
| Related structure data |  7nntMC  7nnuC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10941 (Title: Cryo-EM structure of the folate-specific ECF transporter complex in DDM micelles (200 kV) EMPIAR-10941 (Title: Cryo-EM structure of the folate-specific ECF transporter complex in DDM micelles (200 kV)Data size: 141.7 Data #1: Unaligned movie frames of ECF-FolT2 in DDM micelles (Dataset 1, Quantifoil grid) [micrographs - multiframe] Data #2: Unaligned movie frames of ECF-FolT2 in DDM micelles (Dataset 2, UltrAuFoil grid) [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12483.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12483.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Inward-facing apo conformation of the folate-specific ECF transporter complex in DDM micelles at 3.4 A resolution sharpened at -62 A^2. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.012 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12483_msk_1.map emd_12483_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened map obtained with LocalDeblur used for model...
| File | emd_12483_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map obtained with LocalDeblur used for model building and figures. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1 used during refinement and FSC...
| File | emd_12483_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 used during refinement and FSC gold-standard resolution calculation. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2 used during refinement and FSC...
| File | emd_12483_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 used during refinement and FSC gold-standard resolution calculation. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Folate-specific ECF transporter complex
| Entire | Name: Folate-specific ECF transporter complex |
|---|---|
| Components |
|
-Supramolecule #1: Folate-specific ECF transporter complex
| Supramolecule | Name: Folate-specific ECF transporter complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Lactobacillus delbrueckii subsp. bulgaricus (strain ATCC 11842 / DSM 20081 / JCM 1002 / NBRC 13953 / NCIMB 11778) (bacteria) Lactobacillus delbrueckii subsp. bulgaricus (strain ATCC 11842 / DSM 20081 / JCM 1002 / NBRC 13953 / NCIMB 11778) (bacteria)Strain: ATCC 11842 / DSM 20081 / JCM 1002 / NBRC 13953 / NCIMB 11778 |
-Macromolecule #1: Energy-coupling factor transporter ATP-binding protein EcfA1
| Macromolecule | Name: Energy-coupling factor transporter ATP-binding protein EcfA1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: Translocases |
|---|---|
| Source (natural) | Organism:  Lactobacillus delbrueckii subsp. bulgaricus (strain ATCC 11842 / DSM 20081 / JCM 1002 / NBRC 13953 / NCIMB 11778) (bacteria) Lactobacillus delbrueckii subsp. bulgaricus (strain ATCC 11842 / DSM 20081 / JCM 1002 / NBRC 13953 / NCIMB 11778) (bacteria)Strain: ATCC 11842 / DSM 20081 / JCM 1002 / NBRC 13953 / NCIMB 11778 |
| Molecular weight | Theoretical: 33.166418 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHHHH HGENLYFQGS DNIISFDHVT FTYPDSPRPA LSDLSFAIER GSWTALIGHN GSGKSTVSKL INGLLAPDDL DKSSITVDG VKLGADTVWE VREKVGIVFQ NPDNQFVGAT VSDDVAFGLE NRAVPRPEML KIVAQAVADV GMADYADSEP S NLSGGQKQ ...String: MHHHHHHHHH HGENLYFQGS DNIISFDHVT FTYPDSPRPA LSDLSFAIER GSWTALIGHN GSGKSTVSKL INGLLAPDDL DKSSITVDG VKLGADTVWE VREKVGIVFQ NPDNQFVGAT VSDDVAFGLE NRAVPRPEML KIVAQAVADV GMADYADSEP S NLSGGQKQ RVAIAGILAV KPQVIILDES TSMLDPEGKE QILDLVRKIK EDNNLTVISI THDLEEAAGA DQVLVLDDGQ LL DQGKPEE IFPKVEMLKR IGLDIPFVYR LKQLLKERGI VLPDEIDDDE KLVQSLWQLN SKM UniProtKB: Energy-coupling factor transporter ATP-binding protein EcfA1 |
-Macromolecule #2: Energy-coupling factor transporter ATP-binding protein EcfA2
| Macromolecule | Name: Energy-coupling factor transporter ATP-binding protein EcfA2 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to catalyse transmembrane movement of substances |
|---|---|
| Source (natural) | Organism:  Lactobacillus delbrueckii subsp. bulgaricus (strain ATCC 11842 / DSM 20081 / JCM 1002 / NBRC 13953 / NCIMB 11778) (bacteria) Lactobacillus delbrueckii subsp. bulgaricus (strain ATCC 11842 / DSM 20081 / JCM 1002 / NBRC 13953 / NCIMB 11778) (bacteria)Strain: ATCC 11842 / DSM 20081 / JCM 1002 / NBRC 13953 / NCIMB 11778 |
| Molecular weight | Theoretical: 31.672156 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAIKFENVSY VYSPGSPLEA IGLDQLNFSL EEGKFIALVG HTGSGKSTLM QHFNALLKPT SGKIEIAGYT ITPETGNKGL KDLRRKVSL AFQFSEAQLF ENTVLKDVEY GPRNFGFSED EAREAALKWL KKVGLKDDLI EHSPFDLSGG QMRRVALAGV L AYEPEIIC ...String: MAIKFENVSY VYSPGSPLEA IGLDQLNFSL EEGKFIALVG HTGSGKSTLM QHFNALLKPT SGKIEIAGYT ITPETGNKGL KDLRRKVSL AFQFSEAQLF ENTVLKDVEY GPRNFGFSED EAREAALKWL KKVGLKDDLI EHSPFDLSGG QMRRVALAGV L AYEPEIIC LDEPAAGLDP MGRLEMMQLF KDYQAAGHTV ILVTHNMDDV ADYADDVLAL EHGRLIKHAS PKEVFKDSEW LQ KHHLAEP RSARFAAKLE AAGLKLPGQP LTMPELADAI KQSLKGGEHE UniProtKB: Energy-coupling factor transporter ATP-binding protein EcfA2 |
-Macromolecule #3: Folate family ECF transporter S component
| Macromolecule | Name: Folate family ECF transporter S component / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lactobacillus delbrueckii subsp. bulgaricus (strain ATCC 11842 / DSM 20081 / JCM 1002 / NBRC 13953 / NCIMB 11778) (bacteria) Lactobacillus delbrueckii subsp. bulgaricus (strain ATCC 11842 / DSM 20081 / JCM 1002 / NBRC 13953 / NCIMB 11778) (bacteria)Strain: ATCC 11842 / DSM 20081 / JCM 1002 / NBRC 13953 / NCIMB 11778 |
| Molecular weight | Theoretical: 20.483604 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKSESKVSSK LELRELVLLA MVIAIKVILG QFKVGNATLQ VGLGFIGSVM LGYLFGPWWG FAGGALSDLV SSVIFGNLGG FFIGFTLTA ALGPMIYGFF LYKQPIQIWR VIASVICVTV ICNIGLNTLW VSMMYGINFM VALSSRILKE MITPWIQMVA V WFILEGLS RVKLSRKFWS HPQFEK UniProtKB: Conserved hypothetical membrane protein |
-Macromolecule #4: Energy-coupling factor transporter transmembrane protein EcfT
| Macromolecule | Name: Energy-coupling factor transporter transmembrane protein EcfT type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lactobacillus delbrueckii subsp. bulgaricus (strain ATCC 11842 / DSM 20081 / JCM 1002 / NBRC 13953 / NCIMB 11778) (bacteria) Lactobacillus delbrueckii subsp. bulgaricus (strain ATCC 11842 / DSM 20081 / JCM 1002 / NBRC 13953 / NCIMB 11778) (bacteria)Strain: ATCC 11842 / DSM 20081 / JCM 1002 / NBRC 13953 / NCIMB 11778 |
| Molecular weight | Theoretical: 30.290283 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSKIIIGRYL PGTTFVYRVD PRAKLLTTFY FIIMIFLANN WVSYLVISIF GLAYVFATGL KARVFWDGVK PMIWMIVFTS LLQTFFMAG GKVYWHWWIF TLSSEGLING LYVFIRFAMI ILVSTVMTVT TKPLEIADAM EWMLTPLKLF KVNVGMISLV I SIALRFVP ...String: MSKIIIGRYL PGTTFVYRVD PRAKLLTTFY FIIMIFLANN WVSYLVISIF GLAYVFATGL KARVFWDGVK PMIWMIVFTS LLQTFFMAG GKVYWHWWIF TLSSEGLING LYVFIRFAMI ILVSTVMTVT TKPLEIADAM EWMLTPLKLF KVNVGMISLV I SIALRFVP TLFDQTVKIM NAQRSRGADF NDGGLVKRAK SVVPMLVPLF IDSLEVALDL STAMESRGYK GSEGRTRYRI LE WSKVDLI PVAYCLLLTI LMITTRKH UniProtKB: Energy-coupling factor transporter transmembrane protein EcfT |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 7.9 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 20 mM Tris, pH 8.0, 150 mM NaCl, 0.0261 % (w/v) DDM |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Details: 5 mA; for dataset 2. |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % / Chamber temperature: 288 K / Instrument: FEI VITROBOT MARK IV Details: Grid was blotted for 4 sec after a wait time for 2.5 sec.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-60 / Number grids imaged: 2 / Number real images: 823 / Average exposure time: 9.0 sec. / Average electron dose: 53.3 e/Å2 Details: Dataset 1: 466 movies (Quantifoil grid) Dataset 2: 357 movies (UltrAuFoil grid) |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL | ||||||||||
| Output model |  PDB-7nnt: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)