[English] 日本語
 Yorodumi
Yorodumi- EMDB-12289: Structure of the in situ actomyosin complex from the A-band of mo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12289 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the in situ actomyosin complex from the A-band of mouse psoas muscle sarcomere in the rigor state obtained by sub-tomogram averaging | ||||||||||||||||||
 Map data Map data | Averaged map of in situ actomyosin complex | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Muscle proteins / force generation / sarcomere / cytoskeleton / MOTOR PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of heart rate by epinephrine / muscle thin filament tropomyosin / Formation of the dystrophin-glycoprotein complex (DGC) / Striated Muscle Contraction / Smooth Muscle Contraction / muscle myosin complex / myosin filament / myosin complex / structural constituent of muscle / ventricular cardiac muscle tissue morphogenesis ...positive regulation of heart rate by epinephrine / muscle thin filament tropomyosin / Formation of the dystrophin-glycoprotein complex (DGC) / Striated Muscle Contraction / Smooth Muscle Contraction / muscle myosin complex / myosin filament / myosin complex / structural constituent of muscle / ventricular cardiac muscle tissue morphogenesis / response to muscle activity / mesenchyme migration / myofibril / cytoskeletal motor activity / striated muscle thin filament / skeletal muscle thin filament assembly / skeletal muscle tissue development / skeletal muscle fiber development / cardiac muscle contraction / stress fiber / muscle contraction / actin filament / filopodium / structural constituent of cytoskeleton / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / actin filament binding / actin cytoskeleton / lamellipodium / double-stranded RNA binding / cell body / in utero embryonic development / calmodulin binding / protein heterodimerization activity / hydrolase activity / calcium ion binding / positive regulation of gene expression / protein homodimerization activity / protein-containing complex / ATP binding / identical protein binding / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 10.2 Å | ||||||||||||||||||
 Authors Authors | Wang Z / Grange M | ||||||||||||||||||
| Funding support |  Germany, Germany,  United Kingdom, European Union, 5 items United Kingdom, European Union, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2021 Journal: Cell / Year: 2021Title: The molecular basis for sarcomere organization in vertebrate skeletal muscle. Authors: Zhexin Wang / Michael Grange / Thorsten Wagner / Ay Lin Kho / Mathias Gautel / Stefan Raunser /   Abstract: Sarcomeres are force-generating and load-bearing devices of muscles. A precise molecular picture of how sarcomeres are built underpins understanding their role in health and disease. Here, we ...Sarcomeres are force-generating and load-bearing devices of muscles. A precise molecular picture of how sarcomeres are built underpins understanding their role in health and disease. Here, we determine the molecular architecture of native vertebrate skeletal sarcomeres by electron cryo-tomography. Our reconstruction reveals molecular details of the three-dimensional organization and interaction of actin and myosin in the A-band, I-band, and Z-disc and demonstrates that α-actinin cross-links antiparallel actin filaments by forming doublets with 6-nm spacing. Structures of myosin, tropomyosin, and actin at ~10 Å further reveal two conformations of the "double-head" myosin, where the flexible orientation of the lever arm and light chains enable myosin not only to interact with the same actin filament, but also to split between two actin filaments. Our results provide unexpected insights into the fundamental organization of vertebrate skeletal muscle and serve as a strong foundation for future investigations of muscle diseases. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12289.map.gz emd_12289.map.gz | 4.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12289-v30.xml emd-12289-v30.xml emd-12289.xml emd-12289.xml | 25.7 KB 25.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12289_fsc.xml emd_12289_fsc.xml | 9.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_12289.png emd_12289.png | 102.7 KB | ||
| Masks |  emd_12289_msk_1.map emd_12289_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-12289.cif.gz emd-12289.cif.gz | 7.2 KB | ||
| Others |  emd_12289_half_map_1.map.gz emd_12289_half_map_1.map.gz emd_12289_half_map_2.map.gz emd_12289_half_map_2.map.gz | 23.4 MB 23.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12289 http://ftp.pdbj.org/pub/emdb/structures/EMD-12289 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12289 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12289 | HTTPS FTP |
-Related structure data
| Related structure data |  7nepMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12289.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12289.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Averaged map of in situ actomyosin complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.755 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
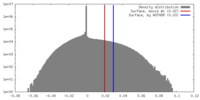
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12289_msk_1.map emd_12289_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
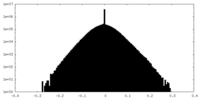
| Density Histograms |
-Half map: Half map 2
| File | emd_12289_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
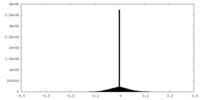
| Density Histograms |
-Half map: Half map 1
| File | emd_12289_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
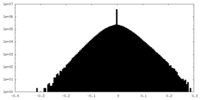
| Density Histograms |
- Sample components
Sample components
-Entire : In situ actomyosin complex in the rigor state from mouse psoas muscle
| Entire | Name: In situ actomyosin complex in the rigor state from mouse psoas muscle |
|---|---|
| Components |
|
-Supramolecule #1: In situ actomyosin complex in the rigor state from mouse psoas muscle
| Supramolecule | Name: In situ actomyosin complex in the rigor state from mouse psoas muscle type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: actin
| Supramolecule | Name: actin / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: myosin double head
| Supramolecule | Name: myosin double head / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#5 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #4: tropomyosin
| Supramolecule | Name: tropomyosin / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Actin, alpha skeletal muscle
| Macromolecule | Name: Actin, alpha skeletal muscle / type: protein_or_peptide / ID: 1 / Number of copies: 11 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.747527 KDa |
| Sequence | String: EDETTALVCD NGSGLVKAGF AGDDAPRAVF PSIVGRPRHQ GVMVGMGQKD SYVGDEAQSK RGILTLKYPI EHGIITNWDD MEKIWHHTF YNELRVAPEE HPTLLTEAPL NPKANREKMT QIMFETFNVP AMYVAIQAVL SLYASGRTTG IVLDSGDGVT H NVPIYEGY ...String: EDETTALVCD NGSGLVKAGF AGDDAPRAVF PSIVGRPRHQ GVMVGMGQKD SYVGDEAQSK RGILTLKYPI EHGIITNWDD MEKIWHHTF YNELRVAPEE HPTLLTEAPL NPKANREKMT QIMFETFNVP AMYVAIQAVL SLYASGRTTG IVLDSGDGVT H NVPIYEGY ALPHAIMRLD LAGRDLTDYL MKILTERGYS FVTTAEREIV RDIKEKLCYV ALDFENEMAT AASSSSLEKS YE LPDGQVI TIGNERFRCP ETLFQPSFIG MESAGIHETT YNSIMKCDID IRKDLYANNV MSGGTTMYPG IADRMQKEIT ALA PSTMKI KIIAPPERKY SVWIGGSILA SLSTFQQMWI TKQEYDEAGP SIVHRKCF UniProtKB: Actin, alpha skeletal muscle |
-Macromolecule #2: Tropomyosin alpha-1 chain
| Macromolecule | Name: Tropomyosin alpha-1 chain / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.922074 KDa |
| Sequence | String: LKLDKENALD RAEQAEADKK AAEDRSKQLE DELVSLQKKL KGTEDELDKY SEALKDAQEK LELAEKKATD AEADVASLNR RIQLVEEEL DRAQERLATA LQKLEEAEKA ADESERGMKV IESRAQKDEE KMEIQEIQLK EAKHIAEDAD RKYEEVARKL V IIESDLER ...String: LKLDKENALD RAEQAEADKK AAEDRSKQLE DELVSLQKKL KGTEDELDKY SEALKDAQEK LELAEKKATD AEADVASLNR RIQLVEEEL DRAQERLATA LQKLEEAEKA ADESERGMKV IESRAQKDEE KMEIQEIQLK EAKHIAEDAD RKYEEVARKL V IIESDLER AEERAELSEG KCAELEEELK TVTNNLKSLE AQAEKYSQKE DKYEEEIKVL SDKLKEAETR AEFAERSVTK LE KSIDDLE DELY UniProtKB: Tropomyosin alpha-1 chain |
-Macromolecule #3: Myosin-4
| Macromolecule | Name: Myosin-4 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 92.518234 KDa |
| Sequence | String: FDAKSSVFVV DAKESYVKAT VQSREGGKVT AKTEGGATVT VKDDQVFSMN PPKYDKIEDM AMMTHLHEPA VLYNLKERYA AWMIYTYSG LFCVTVNPYK WLPVYNPEVV AAYRGKKRQE APPHIFSISD NAYQFMLTDR ENQSILITGE SGAGKTVNTK R VIQYFATI ...String: FDAKSSVFVV DAKESYVKAT VQSREGGKVT AKTEGGATVT VKDDQVFSMN PPKYDKIEDM AMMTHLHEPA VLYNLKERYA AWMIYTYSG LFCVTVNPYK WLPVYNPEVV AAYRGKKRQE APPHIFSISD NAYQFMLTDR ENQSILITGE SGAGKTVNTK R VIQYFATI AVTGDKKKEE ATSGKMQGTL EDQIISANPL LEAFGNAKTV RNDNSSRFGK FIRIHFGATG KLASADIETY LL EKSRVTF QLKAERSYHI FYQIMSNKKP ELIEMLLITT NPYDFAYVSQ GEITVPSIDD QEELMATDTA VDILGFSADE KVA IYKLTG AVMHYGNMKF KQKQREEQAE PDGTEVADKA AYLTSLNSAD LLKALCYPRV KVGNEYVTKG QTVQQVYNSV GALA KSMYE KMFLWMVTRI NQQLDTKQPR QYFIGVLDIA GFEIFDFNTL EQLCINFTNE KLQQFFNHHM FVLEQEEYKK EGIDW EFID FGMDLAACIE LIEKPMGIFS ILEEECMFPK ATDTSFKNKL YEQHLGKSNN FQKPKPAKGK AEAHFSLVHY AGTVDY NII GWLDKNKDPL NETVVGLYQK SGLKTLAFLF SGGQAAEAEG GGGKKGGKKK GSSFQTVSAL FRENLNKLMT NLKSTHP HF VRCLIPNETK TPGAMEHELV LHQLRCNGVL EGIRICRKGF PSRILYADFK QRYKVLNASA IPEGQFIDSK KASEKLLG S IDIDHTQYKF GHTKVFFKAG LLGTLEEMRD EKLAQLITRT QAVCRGYLMR VEFKKMMERR ESIFCIQYNV RAFMNVKHW PWMKLYFKIK PLL UniProtKB: Myosin-4 |
-Macromolecule #4: Myosin light chain 1/3, skeletal muscle isoform
| Macromolecule | Name: Myosin light chain 1/3, skeletal muscle isoform / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.57359 KDa |
| Sequence | String: EFSKEQQEDF KEAFLLFDRT GECKITLSQV GDVLRALGTN PTNAEVKKVL GNPSNEEMNA KKIEFEQFLP MMQAISNNKD QGGYEDFVE GLRVFDKEGN GTVMGAELRH VLATLGEKMK EEEVEALLAG QEDSNGCINY EAFVKHIMS UniProtKB: Myosin light chain 1/3, skeletal muscle isoform |
-Macromolecule #5: Myosin regulatory light chain 2, skeletal muscle isoform
| Macromolecule | Name: Myosin regulatory light chain 2, skeletal muscle isoform type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 16.326439 KDa |
| Sequence | String: MFDQTQIQEF KEAFTVIDQN RDGIIDKEDL RDTFAAMGRL NVKNEELDAM MKEASGPINF TVFLTMFGEK LKGADPEDVI TGAFKVLDP EGKGTIKKQF LEELLTTQCD RFSQEEIKNM WAAFPPDVGG NVDYKNICYV ITHGD UniProtKB: Myosin regulatory light chain 11 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 Component:
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 286 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||||||||
| Details | The sample was myofibrils. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 1-8 / Average electron dose: 3.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 28409 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Refinement | Protocol: RIGID BODY FIT | ||||||||
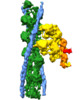
| Output model |  PDB-7nep: |
 Movie
Movie Controller
Controller


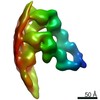


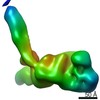












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)