[English] 日本語
 Yorodumi
Yorodumi- EMDB-12242: Bacterial 30S ribosomal subunit assembly complex state E (body domain) -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12242 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bacterial 30S ribosomal subunit assembly complex state E (body domain) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cryo-EM / 30S biogenesis / ribosome assembly / RbfA / RsgA / YjeQ / RimP / KsgA / RsmA / RIBOSOME | |||||||||
| Function / homology |  Function and homology information Function and homology informationornithine decarboxylase inhibitor activity / ribosomal small subunit binding / misfolded RNA binding / Group I intron splicing / RNA folding / four-way junction DNA binding / regulation of mRNA stability / negative regulation of translational initiation / response to cold / mRNA regulatory element binding translation repressor activity ...ornithine decarboxylase inhibitor activity / ribosomal small subunit binding / misfolded RNA binding / Group I intron splicing / RNA folding / four-way junction DNA binding / regulation of mRNA stability / negative regulation of translational initiation / response to cold / mRNA regulatory element binding translation repressor activity / positive regulation of RNA splicing / DNA endonuclease activity / maturation of SSU-rRNA / transcription antitermination / DNA-templated transcription termination / maintenance of translational fidelity / mRNA 5'-UTR binding / ribosome biogenesis / regulation of translation / ribosomal small subunit biogenesis / ribosomal small subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / cytoplasmic translation / tRNA binding / rRNA binding / structural constituent of ribosome / ribosome / translation / response to antibiotic / DNA damage response / zinc ion binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
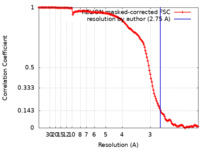
| Method | single particle reconstruction / cryo EM / Resolution: 2.75 Å | |||||||||
 Authors Authors | Schedlbauer A / Iturrioz I | |||||||||
| Funding support |  Spain, 2 items Spain, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2021 Journal: Sci Adv / Year: 2021Title: A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit. Authors: Andreas Schedlbauer / Idoia Iturrioz / Borja Ochoa-Lizarralde / Tammo Diercks / Jorge Pedro López-Alonso / José Luis Lavin / Tatsuya Kaminishi / Retina Çapuni / Neha Dhimole / Elisa de ...Authors: Andreas Schedlbauer / Idoia Iturrioz / Borja Ochoa-Lizarralde / Tammo Diercks / Jorge Pedro López-Alonso / José Luis Lavin / Tatsuya Kaminishi / Retina Çapuni / Neha Dhimole / Elisa de Astigarraga / David Gil-Carton / Paola Fucini / Sean R Connell /   Abstract: While a structural description of the molecular mechanisms guiding ribosome assembly in eukaryotic systems is emerging, bacteria use an unrelated core set of assembly factors for which high- ...While a structural description of the molecular mechanisms guiding ribosome assembly in eukaryotic systems is emerging, bacteria use an unrelated core set of assembly factors for which high-resolution structural information is still missing. To address this, we used single-particle cryo-electron microscopy to visualize the effects of bacterial ribosome assembly factors RimP, RbfA, RsmA, and RsgA on the conformational landscape of the 30 ribosomal subunit and obtained eight snapshots representing late steps in the folding of the decoding center. Analysis of these structures identifies a conserved secondary structure switch in the 16 ribosomal RNA central to decoding site maturation and suggests both a sequential order of action and molecular mechanisms for the assembly factors in coordinating and controlling this switch. Structural and mechanistic parallels between bacterial and eukaryotic systems indicate common folding features inherent to all ribosomes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12242.map.gz emd_12242.map.gz | 182.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12242-v30.xml emd-12242-v30.xml emd-12242.xml emd-12242.xml | 32.6 KB 32.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12242_fsc.xml emd_12242_fsc.xml | 14.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_12242.png emd_12242.png | 169.2 KB | ||
| Masks |  emd_12242_msk_1.map emd_12242_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-12242.cif.gz emd-12242.cif.gz | 8.6 KB | ||
| Others |  emd_12242_half_map_1.map.gz emd_12242_half_map_1.map.gz emd_12242_half_map_2.map.gz emd_12242_half_map_2.map.gz | 183.1 MB 183.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12242 http://ftp.pdbj.org/pub/emdb/structures/EMD-12242 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12242 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12242 | HTTPS FTP |
-Validation report
| Summary document |  emd_12242_validation.pdf.gz emd_12242_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12242_full_validation.pdf.gz emd_12242_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_12242_validation.xml.gz emd_12242_validation.xml.gz | 21 KB | Display | |
| Data in CIF |  emd_12242_validation.cif.gz emd_12242_validation.cif.gz | 27.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12242 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12242 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12242 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12242 | HTTPS FTP |
-Related structure data
| Related structure data |  7bogMC  7af3C  7af5C  7af8C  7afaC  7afdC  7afhC  7afiC  7afkC  7aflC  7afnC  7afoC  7afqC  7afrC  7bodC  7boeC  7bofC  7bohC  7boiC  7narC  7nasC  7natC  7nauC  7navC  7naxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12242.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12242.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12242_msk_1.map emd_12242_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_12242_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_12242_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Bacterial 30S ribosomal subunit assembly complex state E (body domain)
+Supramolecule #1: Bacterial 30S ribosomal subunit assembly complex state E (body domain)
+Supramolecule #2: Bacterial 30S ribosomal subunit assembly
+Supramolecule #3: 30S ribosome-binding factor
+Macromolecule #1: 16S rRNA
+Macromolecule #2: 30S ribosomal protein S4
+Macromolecule #3: 30S ribosomal protein S5
+Macromolecule #4: 30S ribosomal protein S6
+Macromolecule #5: 30S ribosomal protein S8
+Macromolecule #6: 30S ribosomal protein S11
+Macromolecule #7: 30S ribosomal protein S12
+Macromolecule #8: 30S ribosomal protein S15
+Macromolecule #9: 30S ribosomal protein S16
+Macromolecule #10: 30S ribosomal protein S17
+Macromolecule #11: 30S ribosomal protein S18
+Macromolecule #12: 30S ribosomal protein S20
+Macromolecule #13: 30S ribosome-binding factor
+Macromolecule #14: MAGNESIUM ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Support film - Material: CARBON / Support film - topology: CONTINUOUS |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 38.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)