[English] 日本語
 Yorodumi
Yorodumi- EMDB-12191: Cryo-EM structure of the Phosphatidylinositol 3-kinase type 2a (P... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12191 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Phosphatidylinositol 3-kinase type 2a (PI3KC2a) of the class II PI3K family | |||||||||
 Map data Map data | cryosparc refined map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of zinc ion transmembrane transport / Synthesis of PIPs at the early endosome membrane / Synthesis of PIPs at the late endosome membrane / Synthesis of PIPs at the Golgi membrane / Synthesis of PIPs at the plasma membrane / autophagosome organization / Golgi Associated Vesicle Biogenesis / phosphatidylinositol-4-phosphate 3-kinase / Clathrin-mediated endocytosis / 1-phosphatidylinositol-4-phosphate 3-kinase activity ...negative regulation of zinc ion transmembrane transport / Synthesis of PIPs at the early endosome membrane / Synthesis of PIPs at the late endosome membrane / Synthesis of PIPs at the Golgi membrane / Synthesis of PIPs at the plasma membrane / autophagosome organization / Golgi Associated Vesicle Biogenesis / phosphatidylinositol-4-phosphate 3-kinase / Clathrin-mediated endocytosis / 1-phosphatidylinositol-4-phosphate 3-kinase activity / phosphatidylinositol-3-phosphate biosynthetic process / clathrin-coated vesicle / 1-phosphatidylinositol-4,5-bisphosphate 3-kinase activity / phosphatidylinositol-4,5-bisphosphate 3-kinase / phosphatidylinositol 3-kinase / 1-phosphatidylinositol-3-kinase activity / clathrin binding / exocytosis / phosphatidylinositol binding / cellular response to starvation / macroautophagy / trans-Golgi network / endocytosis / vesicle / ATP binding / nucleus / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
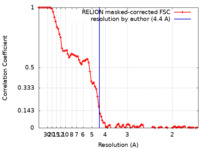
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Lo WT / Zhang Y / Vadas O / Belabed H / Roeske Y / Gulluni F / De Santis MC / Vujicic Zagar A / Stephanowitz H / Hirsch E ...Lo WT / Zhang Y / Vadas O / Belabed H / Roeske Y / Gulluni F / De Santis MC / Vujicic Zagar A / Stephanowitz H / Hirsch E / Liu F / Daumke O / Nazare M / Kudryashev M / Haucke V | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Structural basis of phosphatidylinositol 3-kinase C2α function. Authors: Wen-Ting Lo / Yingyi Zhang / Oscar Vadas / Yvette Roske / Federico Gulluni / Maria Chiara De Santis / Andreja Vujicic Zagar / Heike Stephanowitz / Emilio Hirsch / Fan Liu / Oliver Daumke / ...Authors: Wen-Ting Lo / Yingyi Zhang / Oscar Vadas / Yvette Roske / Federico Gulluni / Maria Chiara De Santis / Andreja Vujicic Zagar / Heike Stephanowitz / Emilio Hirsch / Fan Liu / Oliver Daumke / Misha Kudryashev / Volker Haucke /     Abstract: Phosphatidylinositol 3-kinase type 2α (PI3KC2α) is an essential member of the structurally unresolved class II PI3K family with crucial functions in lipid signaling, endocytosis, angiogenesis, ...Phosphatidylinositol 3-kinase type 2α (PI3KC2α) is an essential member of the structurally unresolved class II PI3K family with crucial functions in lipid signaling, endocytosis, angiogenesis, viral replication, platelet formation and a role in mitosis. The molecular basis of these activities of PI3KC2α is poorly understood. Here, we report high-resolution crystal structures as well as a 4.4-Å cryogenic-electron microscopic (cryo-EM) structure of PI3KC2α in active and inactive conformations. We unravel a coincident mechanism of lipid-induced activation of PI3KC2α at membranes that involves large-scale repositioning of its Ras-binding and lipid-binding distal Phox-homology and C-C2 domains, and can serve as a model for the entire class II PI3K family. Moreover, we describe a PI3KC2α-specific helical bundle domain that underlies its scaffolding function at the mitotic spindle. Our results advance our understanding of PI3K biology and pave the way for the development of specific inhibitors of class II PI3K function with wide applications in biomedicine. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12191.map.gz emd_12191.map.gz | 33.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12191-v30.xml emd-12191-v30.xml emd-12191.xml emd-12191.xml | 21.3 KB 21.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12191_fsc.xml emd_12191_fsc.xml | 9.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_12191.png emd_12191.png | 74.2 KB | ||
| Masks |  emd_12191_msk_1.map emd_12191_msk_1.map | 67 MB |  Mask map Mask map | |
| Others |  emd_12191_additional_1.map.gz emd_12191_additional_1.map.gz emd_12191_half_map_1.map.gz emd_12191_half_map_1.map.gz emd_12191_half_map_2.map.gz emd_12191_half_map_2.map.gz | 62.3 MB 62.3 MB 62.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12191 http://ftp.pdbj.org/pub/emdb/structures/EMD-12191 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12191 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12191 | HTTPS FTP |
-Related structure data
| Related structure data |  7bi2C  7bi4C  7bi6C  7bi9C C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10665 (Title: Single particle Cryo-EM data set for study the structural basis of Phosphatidylinositol 3-kinase type 2α (PI3KC2α) EMPIAR-10665 (Title: Single particle Cryo-EM data set for study the structural basis of Phosphatidylinositol 3-kinase type 2α (PI3KC2α)Data size: 9.5 TB Data #1: -30 degree tilted PI3KC2a data set raw movies [micrographs - multiframe] Data #2: untilted PI3KC2a data set raw movies [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12191.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12191.map.gz / Format: CCP4 / Size: 67 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryosparc refined map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.837 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
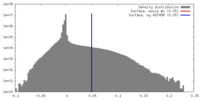
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12191_msk_1.map emd_12191_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: RELION postprocess map
| File | emd_12191_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RELION postprocess map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_12191_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_12191_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Class II PI3KA
| Entire | Name: Class II PI3KA |
|---|---|
| Components |
|
-Supramolecule #1: Class II PI3KA
| Supramolecule | Name: Class II PI3KA / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Experimental: 150 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.8 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
Details: The buffer containing 20 mM Tris-HCl, 100 mM NaCl at pH 7.4. | |||||||||
| Grid | Model: Quantifoil / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: OTHER Details: QUANTIFOIL Holey Au-carbon-R2/2 specimen grids were used. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV Details: The sample was vitrified by plunge-freezing into liquid ethane using a Mark IV Vitrobot device (Thermo Fisher Scientific), blotting force 20, blotting time 4.5-5.5s.. | |||||||||
| Details | The purified sample was diluted to 0.8 mg/ml into buffer containing 20 mM Tris-HCl, 100 mM NaCl at pH 7.4. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 79.8 K / Max: 89.9 K |
| Specialist optics | Spherical aberration corrector: no / Chromatic aberration corrector: no / Energy filter - Slit width: 20 eV |
| Details | Grids were screened first to check ice thickness and particle distribution. |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 2 / Number real images: 10433 / Average exposure time: 3.0 sec. / Average electron dose: 60.0 e/Å2 Details: Images were collected with 50 frames over 60 electrons per squared angstrom. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 3.3000000000000003 µm / Calibrated defocus min: 1.2 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)