+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 7bi2 | ||||||
|---|---|---|---|---|---|---|---|
| Title | PI3KC2aDeltaN and DeltaC-C2 | ||||||
 Components Components | Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha,Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha | ||||||
 Keywords Keywords | TRANSFERASE / PI3KC2 alpha / kinase | ||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of zinc ion transmembrane transport / Synthesis of PIPs at the early endosome membrane / Synthesis of PIPs at the late endosome membrane / Synthesis of PIPs at the Golgi membrane / Synthesis of PIPs at the plasma membrane / autophagosome organization / Golgi Associated Vesicle Biogenesis / phosphatidylinositol-4-phosphate 3-kinase / Clathrin-mediated endocytosis / 1-phosphatidylinositol-4-phosphate 3-kinase activity ...negative regulation of zinc ion transmembrane transport / Synthesis of PIPs at the early endosome membrane / Synthesis of PIPs at the late endosome membrane / Synthesis of PIPs at the Golgi membrane / Synthesis of PIPs at the plasma membrane / autophagosome organization / Golgi Associated Vesicle Biogenesis / phosphatidylinositol-4-phosphate 3-kinase / Clathrin-mediated endocytosis / 1-phosphatidylinositol-4-phosphate 3-kinase activity / phosphatidylinositol-3-phosphate biosynthetic process / clathrin-coated vesicle / 1-phosphatidylinositol-4,5-bisphosphate 3-kinase activity / phosphatidylinositol-4,5-bisphosphate 3-kinase / phosphatidylinositol 3-kinase / 1-phosphatidylinositol-3-kinase activity / clathrin binding / exocytosis / phosphatidylinositol binding / cellular response to starvation / macroautophagy / trans-Golgi network / endocytosis / vesicle / ATP binding / nucleus / plasma membrane / cytoplasm Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 3.25 Å MOLECULAR REPLACEMENT / Resolution: 3.25 Å | ||||||
 Authors Authors | Lo, W.T. / Roske, Y. / Daumke, O. / Haucke, V. | ||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2022 Journal: Nat Struct Mol Biol / Year: 2022Title: Structural basis of phosphatidylinositol 3-kinase C2α function. Authors: Wen-Ting Lo / Yingyi Zhang / Oscar Vadas / Yvette Roske / Federico Gulluni / Maria Chiara De Santis / Andreja Vujicic Zagar / Heike Stephanowitz / Emilio Hirsch / Fan Liu / Oliver Daumke / ...Authors: Wen-Ting Lo / Yingyi Zhang / Oscar Vadas / Yvette Roske / Federico Gulluni / Maria Chiara De Santis / Andreja Vujicic Zagar / Heike Stephanowitz / Emilio Hirsch / Fan Liu / Oliver Daumke / Misha Kudryashev / Volker Haucke /     Abstract: Phosphatidylinositol 3-kinase type 2α (PI3KC2α) is an essential member of the structurally unresolved class II PI3K family with crucial functions in lipid signaling, endocytosis, angiogenesis, ...Phosphatidylinositol 3-kinase type 2α (PI3KC2α) is an essential member of the structurally unresolved class II PI3K family with crucial functions in lipid signaling, endocytosis, angiogenesis, viral replication, platelet formation and a role in mitosis. The molecular basis of these activities of PI3KC2α is poorly understood. Here, we report high-resolution crystal structures as well as a 4.4-Å cryogenic-electron microscopic (cryo-EM) structure of PI3KC2α in active and inactive conformations. We unravel a coincident mechanism of lipid-induced activation of PI3KC2α at membranes that involves large-scale repositioning of its Ras-binding and lipid-binding distal Phox-homology and C-C2 domains, and can serve as a model for the entire class II PI3K family. Moreover, we describe a PI3KC2α-specific helical bundle domain that underlies its scaffolding function at the mitotic spindle. Our results advance our understanding of PI3K biology and pave the way for the development of specific inhibitors of class II PI3K function with wide applications in biomedicine. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  7bi2.cif.gz 7bi2.cif.gz | 431.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb7bi2.ent.gz pdb7bi2.ent.gz | 355.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  7bi2.json.gz 7bi2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bi/7bi2 https://data.pdbj.org/pub/pdb/validation_reports/bi/7bi2 ftp://data.pdbj.org/pub/pdb/validation_reports/bi/7bi2 ftp://data.pdbj.org/pub/pdb/validation_reports/bi/7bi2 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  7bi4C  7bi6C  7bi9C  1e8xS  2iwlS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 131361.906 Da / Num. of mol.: 1 / Mutation: 533-544/GSGS Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: Q61194, phosphatidylinositol-4-phosphate 3-kinase | ||||
|---|---|---|---|---|---|
| #2: Chemical | ChemComp-EDO / | ||||
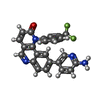
| #3: Chemical | ChemComp-17G / | ||||
| #4: Chemical | ChemComp-IOD / #5: Water | ChemComp-HOH / | Has ligand of interest | N | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.63 Å3/Da / Density % sol: 53.21 % |
|---|---|
| Crystal grow | Temperature: 298 K / Method: vapor diffusion, sitting drop / pH: 7.5 Details: 0.1MTris, 8% PEG20000, 10% ethylene glycol, 10% formamide |
-Data collection
| Diffraction | Mean temperature: 100 K / Serial crystal experiment: N |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  BESSY BESSY  / Beamline: 14.1 / Wavelength: 0.9184 Å / Beamline: 14.1 / Wavelength: 0.9184 Å |
| Detector | Type: DECTRIS PILATUS3 6M / Detector: PIXEL / Date: May 12, 2019 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 0.9184 Å / Relative weight: 1 |
| Reflection | Resolution: 3.25→49.2 Å / Num. obs: 22257 / % possible obs: 99.1 % / Redundancy: 4.9 % / CC1/2: 0.998 / Rrim(I) all: 0.152 / Net I/σ(I): 9.73 |
| Reflection shell | Resolution: 3.25→3.45 Å / Redundancy: 4.9 % / Mean I/σ(I) obs: 0.84 / Num. unique obs: 3455 / CC1/2: 0.443 / Rrim(I) all: 2.294 / % possible all: 97.1 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1E8X, 2IWL Resolution: 3.25→49.2 Å / Cor.coef. Fo:Fc: 0.923 / Cor.coef. Fo:Fc free: 0.889 / SU B: 110.025 / SU ML: 0.748 / Cross valid method: THROUGHOUT / σ(F): 0 / ESU R Free: 0.647 / Stereochemistry target values: MAXIMUM LIKELIHOOD Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS U VALUES : WITH TLS ADDED
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 545.73 Å2 / Biso mean: 137.09 Å2 / Biso min: 72.4 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: final / Resolution: 3.25→49.2 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 3.25→3.334 Å / Rfactor Rfree error: 0 / Total num. of bins used: 20
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller












 PDBj
PDBj