[English] 日本語
 Yorodumi
Yorodumi- EMDB-12138: Cryo-EM structure of fatty acid synthase (FAS) from Pichia pastoris -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12138 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of fatty acid synthase (FAS) from Pichia pastoris | ||||||||||||
 Map data Map data | Map filtered according to local resolution. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Multienzyme / Complex / Fatty acid / Synthase / BIOSYNTHETIC PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationmitotic nuclear membrane biogenesis / palmitic acid biosynthetic process / fatty-acyl-CoA synthase system / fatty acid synthase complex / fatty-acyl-CoA synthase activity / [acyl-carrier-protein] S-acetyltransferase / [acyl-carrier-protein] S-acetyltransferase activity / holo-[acyl-carrier-protein] synthase activity / [acyl-carrier-protein] S-malonyltransferase / [acyl-carrier-protein] S-malonyltransferase activity ...mitotic nuclear membrane biogenesis / palmitic acid biosynthetic process / fatty-acyl-CoA synthase system / fatty acid synthase complex / fatty-acyl-CoA synthase activity / [acyl-carrier-protein] S-acetyltransferase / [acyl-carrier-protein] S-acetyltransferase activity / holo-[acyl-carrier-protein] synthase activity / [acyl-carrier-protein] S-malonyltransferase / [acyl-carrier-protein] S-malonyltransferase activity / 3-hydroxyacyl-[acyl-carrier-protein] dehydratase / beta-ketoacyl-[acyl-carrier-protein] synthase I / (3R)-hydroxyacyl-[acyl-carrier-protein] dehydratase activity / 3-oxoacyl-[acyl-carrier-protein] reductase / 3-oxoacyl-[acyl-carrier-protein] reductase (NADPH) activity / oleoyl-[acyl-carrier-protein] hydrolase / fatty acyl-[ACP] hydrolase activity / enoyl-[acyl-carrier-protein] reductase (NADH) / enoyl-[acyl-carrier-protein] reductase (NADH) activity / fatty acid synthase activity / 3-oxoacyl-[acyl-carrier-protein] synthase activity / fatty acid biosynthetic process / magnesium ion binding Similarity search - Function | ||||||||||||
| Biological species |  Komagataella phaffii GS115 (fungus) / Komagataella phaffii GS115 (fungus) /  Komagataella phaffii (strain GS115 / ATCC 20864) (fungus) Komagataella phaffii (strain GS115 / ATCC 20864) (fungus) | ||||||||||||
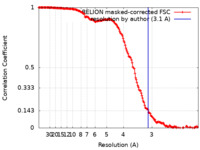
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | ||||||||||||
 Authors Authors | Snowden JS / Alzahrani J | ||||||||||||
| Funding support |  United Kingdom, United Kingdom,  Saudi Arabia, 3 items Saudi Arabia, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2021 Journal: Sci Rep / Year: 2021Title: Structural insight into Pichia pastoris fatty acid synthase. Authors: Joseph S Snowden / Jehad Alzahrani / Lee Sherry / Martin Stacey / David J Rowlands / Neil A Ranson / Nicola J Stonehouse /  Abstract: Type I fatty acid synthases (FASs) are critical metabolic enzymes which are common targets for bioengineering in the production of biofuels and other products. Serendipitously, we identified FAS as a ...Type I fatty acid synthases (FASs) are critical metabolic enzymes which are common targets for bioengineering in the production of biofuels and other products. Serendipitously, we identified FAS as a contaminant in a cryoEM dataset of virus-like particles (VLPs) purified from P. pastoris, an important model organism and common expression system used in protein production. From these data, we determined the structure of P. pastoris FAS to 3.1 Å resolution. While the overall organisation of the complex was typical of type I FASs, we identified several differences in both structural and enzymatic domains through comparison with the prototypical yeast FAS from S. cerevisiae. Using focussed classification, we were also able to resolve and model the mobile acyl-carrier protein (ACP) domain, which is key for function. Ultimately, the structure reported here will be a useful resource for further efforts to engineer yeast FAS for synthesis of alternate products. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12138.map.gz emd_12138.map.gz | 211.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12138-v30.xml emd-12138-v30.xml emd-12138.xml emd-12138.xml | 33 KB 33 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12138_fsc.xml emd_12138_fsc.xml | 16.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_12138.png emd_12138.png | 216.3 KB | ||
| Masks |  emd_12138_msk_1.map emd_12138_msk_1.map | 371.3 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-12138.cif.gz emd-12138.cif.gz | 8.8 KB | ||
| Others |  emd_12138_additional_1.map.gz emd_12138_additional_1.map.gz emd_12138_additional_2.map.gz emd_12138_additional_2.map.gz emd_12138_additional_3.map.gz emd_12138_additional_3.map.gz emd_12138_additional_4.map.gz emd_12138_additional_4.map.gz emd_12138_half_map_1.map.gz emd_12138_half_map_1.map.gz emd_12138_half_map_2.map.gz emd_12138_half_map_2.map.gz | 295.8 MB 186 MB 348.6 MB 48 MB 296.9 MB 298.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12138 http://ftp.pdbj.org/pub/emdb/structures/EMD-12138 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12138 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12138 | HTTPS FTP |
-Validation report
| Summary document |  emd_12138_validation.pdf.gz emd_12138_validation.pdf.gz | 918.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12138_full_validation.pdf.gz emd_12138_full_validation.pdf.gz | 918.5 KB | Display | |
| Data in XML |  emd_12138_validation.xml.gz emd_12138_validation.xml.gz | 23.8 KB | Display | |
| Data in CIF |  emd_12138_validation.cif.gz emd_12138_validation.cif.gz | 31.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12138 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12138 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12138 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12138 | HTTPS FTP |
-Related structure data
| Related structure data |  7bc4MC  7bc5C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12138.map.gz / Format: CCP4 / Size: 371.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12138.map.gz / Format: CCP4 / Size: 371.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map filtered according to local resolution. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
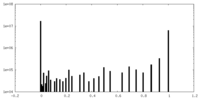
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.065 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
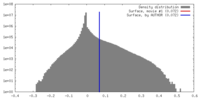
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12138_msk_1.map emd_12138_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
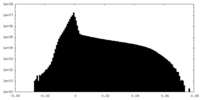
| Projections & Slices |
| ||||||||||||
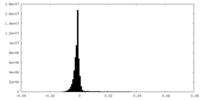
| Density Histograms |
-Additional map: Unsharpened map.
| File | emd_12138_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Map of local resolution values, for local resolution...
| File | emd_12138_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of local resolution values, for local resolution colouring of maps. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened map without solvent mask applied.
| File | emd_12138_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map without solvent mask applied. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Sharpened map with solvent mask applied.
| File | emd_12138_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map with solvent mask applied. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1.
| File | emd_12138_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2.
| File | emd_12138_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Fatty acid synthase
| Entire | Name: Fatty acid synthase |
|---|---|
| Components |
|
-Supramolecule #1: Fatty acid synthase
| Supramolecule | Name: Fatty acid synthase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Komagataella phaffii GS115 (fungus) Komagataella phaffii GS115 (fungus) |
| Molecular weight | Theoretical: 2.6 MDa |
-Macromolecule #1: Fatty acid synthase subunit alpha
| Macromolecule | Name: Fatty acid synthase subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: fatty-acyl-CoA synthase system |
|---|---|
| Source (natural) | Organism:  Komagataella phaffii (strain GS115 / ATCC 20864) (fungus) Komagataella phaffii (strain GS115 / ATCC 20864) (fungus)Strain: GS115 / ATCC 20864 |
| Molecular weight | Theoretical: 206.375734 KDa |
| Sequence | String: MRPEVEQELS HVLLTELLAY QFASPVRWIE TQDVFLKDYN TERVVEIGPS PTLAGMASRT IKAKYESYDA ALSLQRQVLC YAKDTKEIY YTPDPADIAP PIKEEAETSA AATSSSAPAA AAPVSAAPAA APSGPVAEIP DEPVKAALVL HVLVAHKLKK S LDAVPLSK ...String: MRPEVEQELS HVLLTELLAY QFASPVRWIE TQDVFLKDYN TERVVEIGPS PTLAGMASRT IKAKYESYDA ALSLQRQVLC YAKDTKEIY YTPDPADIAP PIKEEAETSA AATSSSAPAA AAPVSAAPAA APSGPVAEIP DEPVKAALVL HVLVAHKLKK S LDAVPLSK AIKDLVGGKS TVQNEILGDL GKEFGSTPEK PEDTPLQELA EQFQDTFPGS LGKQTGSLVN RLMSSKMPGG FS LSVARKY LQTRWGLGPG RQDSVLLVAL VNEPGARLSS DGEAKEFLDS CAQKYASGAG ITLAQAAAGG AGSSGAGGAV IDA EAFEEL TKDNRVLARQ QLEVLARYLK YDLTKGEKSL VKEKEASSLL QQELDLWAEE HGEIYAQGIK PVFSHLKART YDSY WNWAR QDALSMYFDI IFGKLTDVDR ETVSQCIQLM NRSNPTLIKF MQYHIDHCPE YKGETYQLAK SLGQQLIDNC IQVAN QDPV YKDISYPTGP HTEVDSKGNI VYKEVNRKSV RKLEQYVFEM SQGGELTKEV QPTIQEDLAK IYEALNKQAS TESQLE FNK LYNSLIEFVE KSKEIEVSKS INAVLASKSS DSDRSAEISS LSEKTSIVDP VSGGIPPETV PFLHLKKKLP SGEWVFD RD TSALFLDGLQ KGAVNGISYK GKNVLITGAG AGSIGAEVLQ GLISGGAKVI VTTSRFSKKV TEYYQDIYAR FGAAGSCL I VVPFNQGSKQ DVEALIDYIY RDVKDEGLGW DLDAVIPFAA IPEAGIEIDE LGSKSELAHR IMLTNLLRLL GEVKKQKFT RAINTRPAQI ILPLSPNHGT FGSDGLYSES KLGLETLFNR WYSESWSEQL TVCGAIIGWT RGTGLMSGNN IIAEGLEKLG VRTFSQKEM AFNILGLMTP ELTEMCQNGP VVADLNGGLQ FIENLREYTA QLRNEIYETS EVRRAVSIET GIETRVVNGE N ADAPYQKA RIEPRANLKF EFPPLKSHKE IQNKAPGLEG LLDLERVIVV TGFGEVSPWG NTRTRWEMEA FGEFSIEGCL EM AWIMGFI KYHNGNLKGK PYTGWIDAKT NEPVEDKDIK KKYEEEILAH AGIRLIEPEL FRGYNPEKKE LIQEVIIEQD MAP FVTDES TAQQYKLQHE DAVDILKSEE SDEYTVTFKK GARLFVPKAL RFDRLVAGQI PTGWDAKRYG ISEDTISQVD PVTL YALVS TIEALLSAGI TDPYEFYKYV HVSEVGNCSG SGMGGVSALR GMFRDRYSDK PVQNDILQES FINTMSAWVN MLLLS SSGP IKTPVGACAT AVESVDIGVE TILSGKAKIC LVGGYDDFQE EGSYEFANMN ATSNSLDEFD HGRTPQEMSR PATTTR NGF MEAQGSGTQV IMNAELAIKM GVPIYAIVAL TATATDKIGR SVPAPGKGIL TTAREHHGSL KTKSPKLDIK YRTRQLN KR KDQIKQWVED ELEYIREEAA ELANSDAKFD AVSFVSERTE EVYREATKQV KMAQQEWGNE FWKNDPRIAP LRGALATF N LTVDDLGVAS FHGTSTKAND KNESITINKM MQHLGRSEGN PVFGVFQKYL TGHPKGAAGA WMLNGAIQIL QTGIVPGNR NADNVDKILE DFEYVLYPSR SIQTDGIKAC SVTSFGFGQK GGQAIVVHPD YLFASLDSET FEEYKTKVEA RYKSTYRYMH NAIIRNTMF VAKSDPPYTD ELEQPVYLDP LARVNNCKKN PSKLVFVNAD VQSKQNFVGK SANDTAKVIS SLTSDVTSGG K GVGVDVEL ISAINNENHT FIERNFTENE ISYCASAPSS KSSLAGTWSA KEAVFKALGV ESKGAGASLK DIEIVRDSKG AP TVVLHGD AKSAASAAGV KNVKVSISHD DVQSVAVAIS EF UniProtKB: Fatty acid synthase subunit alpha |
-Macromolecule #2: Fatty acid synthase subunit beta
| Macromolecule | Name: Fatty acid synthase subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: fatty-acyl-CoA synthase system |
|---|---|
| Source (natural) | Organism:  Komagataella phaffii (strain GS115 / ATCC 20864) (fungus) Komagataella phaffii (strain GS115 / ATCC 20864) (fungus)Strain: GS115 / ATCC 20864 |
| Molecular weight | Theoretical: 230.000625 KDa |
| Sequence | String: MSATSGVVNR PLVLNHGSIE STILIPTTEY HFYQTLLEGF RKSLPQVTEG FADDDEPSSK AELLMKFLGY IVQSGVSNQQ EQLAAAKLV LNEFESRFLQ GLNLHSYAAI LLKSETFPTT LLKIKENLIK NYYLGRALVY LPGQRGLVYP PSALLNAGKS G SAQIYAIF ...String: MSATSGVVNR PLVLNHGSIE STILIPTTEY HFYQTLLEGF RKSLPQVTEG FADDDEPSSK AELLMKFLGY IVQSGVSNQQ EQLAAAKLV LNEFESRFLQ GLNLHSYAAI LLKSETFPTT LLKIKENLIK NYYLGRALVY LPGQRGLVYP PSALLNAGKS G SAQIYAIF GGQGNTDDYF EELRDIYHIY QGLVSDFVTK AQLKLQELIR TTPETDRIYT QGLDLINWLE NKDKTPDQQQ LL SIPMSCP LICVIQLCHY IVTCRILGIT PGQLRDSLKG TTGHSQGLVT AVVVSSADSW ESFEKLALQA VEFMFYIGVR GLQ TYPNTS LPPSIVQDSE ENAEGTPSPM LSVRDLSYDQ LVKFVNETNQ HLPEAKHIDI SLINGPRNVV LTGPPQSLYG LNLN LRKAK APSGLDQARI PFSERKLRFS NRFLPIMSPF HSHLLSPSTE KIVADLKKAG VEFSQSSMKL PVFDTYDGKD LRSYS GSIA ARLVECITKL RVNWELSTEF NSTHVLDFGP GGASGLGVLT HRNKEGTGSR VIVAGVLDAE SEDSEFGYKQ EIFESN EKA IKYAPNWLKE YKPKLVKTSA GKIFVDTKFS RLLGRAPLMV PGMTPTTVSP DFVAATLNAG FHTEIAGGGY FAPSIMK AA LQRVIDQVTP GTGVGINLIY VNPRMLQWGI PMIKELREQG FPIQSLSIGA GVPSLEVATE YIETLGLAHL GLKPGSID A VNQVITIAKA HPNFPIVLQW TGGRGGGHHS FEDFHQPILQ MYSKIRKCKN IILIAGSGFG SAEDTYPYLT GSWSHQFSY PSMPFDGVLF GSRVMTAKEA KTSPAAKQAI ADCTGVDNSQ WENTYKKPTG GIITVRSEMG EPIHKIATRG VMLWKELDDT IFTLPKNKM LEAIAKKKDY IIKKLNADYQ KPWFAKNEKG TCDLEDMTYK QIAERLVELM YVRKSQRWID VTLRNFTGKF L RRIEERFA TKVGTISLIQ NFSQLEEPEK AIDSVFKAYP EAASQLINEE DCDWFLLEAQ SPTQKPVPFI PVLDERFEFF FK KDSLWQS EDLEAVVGED VQRTCILHGP VAAQFSNKVD EPIKDILENI HKGHIKSLVK EVYNGDESKI PVVEYFSSVD SFS DTAIEG VKIERSRNTE TFTVTSGNVD NQQWFDLLAG KELSWRRAFI TAARLVQGTN FVSNPAHSVL APSKDLVVKI ENGS DAKKT VLTAFQRVRG KYVPAVSLKS IGDLKIKLEL IETRTADKSA VALELFYNYK PTDGFAPILE VMEGRNTSIK NFYWK LWFG SSVPVDLDFD ANKPISGGEA SVSSQAIAEF THAVGNSCED FVPRAGRPQL APMDFAIVLG WKAIMKAIFP KTVDGD ILK LVHLSNGYKM FPGADPLKKG DVVSTVAHIR SVVNGETGKT VEVVGVISRD GKPVLEVNSQ FFYRGKYQDF GNSFKKT TE TPVQVAFKSA KDIAVLKSKE WFHLEKDIDL LNQTLTFRCE SYVKFKSSTV FASVKTTGQA LLELPSKEII QVAEINYE S GSSYGNPVLD YLTRHGSTIE QPIMFENAIP LAQGTELTSK APGTNETYAA VSGDYNPIHV NKVFASYANL PGTITHGMY SSAAVRALVE QWAAQNVATR VRAFKADFVG MVLPNDELVT HLEHVGMING RKIIKVETKK VETEEVVLIG EAEIEQPVST FVFTGQGSQ EQGMGMDLYN SSEVAKSVWD RADVHFINNY GFSILDIVKN NPTELTVHFG GAKGRSIRNN YISMMFETVA A DGQLKSEK IFKEINEDTI SFTFKSPTGL LSATQFTQPA LTLMEKASFE DMKSKGLVPS ESMFAGHSLG EYSALTSLGD VM PIESLVD VVFYRGMTMQ VAVPRDEQGR SNYGMIAVNP SRVSSTFNDS ALRFVVEHIA QQTGWLLEIV NYNVENTQYV AAG DLRGLD TLSNVLNVFK IQKIDIVKLQ ETISLDEVKV HLSEIVDEVS KKSSSKPQPI DLERGFACIP LKGISVPFHS SYLR SGVKP FQTFLCKKIP KSAVKPANLI GKYIPNLTAK PFQLTKEYFE DVYELTKSEK IKHILDHWEE YESS UniProtKB: Fatty acid synthase subunit beta |
-Macromolecule #3: FLAVIN MONONUCLEOTIDE
| Macromolecule | Name: FLAVIN MONONUCLEOTIDE / type: ligand / ID: 3 / Number of copies: 1 / Formula: FMN |
|---|---|
| Molecular weight | Theoretical: 456.344 Da |
| Chemical component information |  ChemComp-FMN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: PBS |
|---|---|
| Grid | Material: COPPER / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 281 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number real images: 3643 / Average electron dose: 60.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)