+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11921 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cytochrome c oxidase structure in P-state | |||||||||
 Map data Map data | density modified map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Terminal oxidase Cytochrome c oxidase aa3 oxidase / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationaerobic electron transport chain / respiratory chain complex IV / cytochrome-c oxidase / oxidative phosphorylation / cytochrome-c oxidase activity / electron transport coupled proton transport / ATP synthesis coupled electron transport / respiratory electron transport chain / copper ion binding / heme binding ...aerobic electron transport chain / respiratory chain complex IV / cytochrome-c oxidase / oxidative phosphorylation / cytochrome-c oxidase activity / electron transport coupled proton transport / ATP synthesis coupled electron transport / respiratory electron transport chain / copper ion binding / heme binding / metal ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Paracoccus denitrificans (bacteria) Paracoccus denitrificans (bacteria) | |||||||||
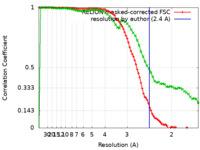
| Method | single particle reconstruction / cryo EM / Resolution: 2.4 Å | |||||||||
 Authors Authors | Kolbe F / Safarian S | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Cryo-EM structures of intermediates suggest an alternative catalytic reaction cycle for cytochrome c oxidase. Authors: F Kolbe / S Safarian / Ż Piórek / S Welsch / H Müller / H Michel /  Abstract: Cytochrome c oxidases are among the most important and fundamental enzymes of life. Integrated into membranes they use four electrons from cytochrome c molecules to reduce molecular oxygen (dioxygen) ...Cytochrome c oxidases are among the most important and fundamental enzymes of life. Integrated into membranes they use four electrons from cytochrome c molecules to reduce molecular oxygen (dioxygen) to water. Their catalytic cycle has been considered to start with the oxidized form. Subsequent electron transfers lead to the E-state, the R-state (which binds oxygen), the P-state (with an already split dioxygen bond), the F-state and the O-state again. Here, we determined structures of up to 1.9 Å resolution of these intermediates by single particle cryo-EM. Our results suggest that in the O-state the active site contains a peroxide dianion and in the P-state possibly an intact dioxygen molecule, the F-state may contain a superoxide anion. Thus, the enzyme's catalytic cycle may have to be turned by 180 degrees. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11921.map.gz emd_11921.map.gz | 10.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11921-v30.xml emd-11921-v30.xml emd-11921.xml emd-11921.xml | 27.1 KB 27.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11921_fsc.xml emd_11921_fsc.xml emd_11921_fsc_2.xml emd_11921_fsc_2.xml | 9.1 KB 7.4 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_11921.png emd_11921.png | 268.9 KB | ||
| Filedesc metadata |  emd-11921.cif.gz emd-11921.cif.gz | 7.4 KB | ||
| Others |  emd_11921_additional_1.map.gz emd_11921_additional_1.map.gz emd_11921_half_map_1.map.gz emd_11921_half_map_1.map.gz emd_11921_half_map_2.map.gz emd_11921_half_map_2.map.gz | 42.9 MB 49.6 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11921 http://ftp.pdbj.org/pub/emdb/structures/EMD-11921 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11921 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11921 | HTTPS FTP |
-Related structure data
| Related structure data |  7ateMC  7atnC  7au3C  7au6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11921.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11921.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | density modified map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.833 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
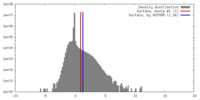
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: relion post process map
| File | emd_11921_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | relion post process map | ||||||||||||
| Projections & Slices |
| ||||||||||||
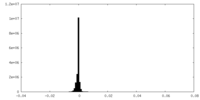
| Density Histograms |
-Half map: relion half map 2
| File | emd_11921_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | relion half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: relion half map 1
| File | emd_11921_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | relion half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Cytochrome c oxidase with four subunits reconstituted in lipid na...
+Supramolecule #1: Cytochrome c oxidase with four subunits reconstituted in lipid na...
+Macromolecule #1: Cytochrome c oxidase subunit 1-beta
+Macromolecule #2: Cytochrome c oxidase subunit 2
+Macromolecule #3: Cytochrome c oxidase subunit 3
+Macromolecule #4: Cytochrome c oxidase subunit 4
+Macromolecule #5: MANGANESE (II) ION
+Macromolecule #6: HEME-A
+Macromolecule #7: COPPER (II) ION
+Macromolecule #8: CALCIUM ION
+Macromolecule #9: HYDROGEN PEROXIDE
+Macromolecule #10: DINUCLEAR COPPER ION
+Macromolecule #11: 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE
+Macromolecule #12: (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-...
+Macromolecule #13: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 8 / Component - Concentration: 50.0 mM / Component - Formula: KPi / Component - Name: Potassium Phosphate |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 4 second before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average exposure time: 30.0 sec. / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)