[English] 日本語
 Yorodumi
Yorodumi- EMDB-11233: Helicobacter pylori urease with inhibitor bound in the active site -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11233 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
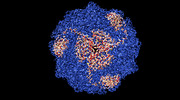
| Title | Helicobacter pylori urease with inhibitor bound in the active site | |||||||||
 Map data Map data | Relion unmasked map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | dodecamer / bi nickel center / enzyme / cytoplasm / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationurease complex / urease / urease activity / urea catabolic process / nickel cation binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
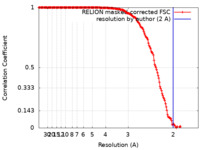
| Method | single particle reconstruction / cryo EM / Resolution: 2.0 Å | |||||||||
 Authors Authors | Luecke H / Cunha E | |||||||||
| Funding support |  Norway, Norway,  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Cryo-EM structure of Helicobacter pylori urease with an inhibitor in the active site at 2.0 Å resolution. Authors: Eva S Cunha / Xiaorui Chen / Marta Sanz-Gaitero / Deryck J Mills / Hartmut Luecke /     Abstract: Infection of the human stomach by Helicobacter pylori remains a worldwide problem and greatly contributes to peptic ulcer disease and gastric cancer. Without active intervention approximately 50% of ...Infection of the human stomach by Helicobacter pylori remains a worldwide problem and greatly contributes to peptic ulcer disease and gastric cancer. Without active intervention approximately 50% of the world population will continue to be infected with this gastric pathogen. Current eradication, called triple therapy, entails a proton-pump inhibitor and two broadband antibiotics, however resistance to either clarithromycin or metronidazole is greater than 25% and rising. Therefore, there is an urgent need for a targeted, high-specificity eradication drug. Gastric infection by H. pylori depends on the expression of a nickel-dependent urease in the cytoplasm of the bacteria. Here, we report the 2.0 Å resolution structure of the 1.1 MDa urease in complex with an inhibitor by cryo-electron microscopy and compare it to a β-mercaptoethanol-inhibited structure at 2.5 Å resolution. The structural information is of sufficient detail to aid in the development of inhibitors with high specificity and affinity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_11233.map.gz emd_11233.map.gz | 370.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-11233-v30.xml emd-11233-v30.xml emd-11233.xml emd-11233.xml | 26.5 KB 26.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_11233_fsc.xml emd_11233_fsc.xml | 16.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_11233.png emd_11233.png | 274 KB | ||
| Masks |  emd_11233_msk_1.map emd_11233_msk_1.map | 396.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-11233.cif.gz emd-11233.cif.gz | 8 KB | ||
| Others |  emd_11233_additional_1.map.gz emd_11233_additional_1.map.gz emd_11233_half_map_1.map.gz emd_11233_half_map_1.map.gz emd_11233_half_map_2.map.gz emd_11233_half_map_2.map.gz | 70.9 MB 314.8 MB 314.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-11233 http://ftp.pdbj.org/pub/emdb/structures/EMD-11233 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11233 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-11233 | HTTPS FTP |
-Related structure data
| Related structure data |  6zjaMC  4629C  6qsuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11233.map.gz / Format: CCP4 / Size: 396.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11233.map.gz / Format: CCP4 / Size: 396.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion unmasked map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8426 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_11233_msk_1.map emd_11233_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Density modified map
| File | emd_11233_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density modified map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 2
| File | emd_11233_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map 1
| File | emd_11233_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : 1.1 MDa Helicobacter pylori Urease
| Entire | Name: 1.1 MDa Helicobacter pylori Urease |
|---|---|
| Components |
|
-Supramolecule #1: 1.1 MDa Helicobacter pylori Urease
| Supramolecule | Name: 1.1 MDa Helicobacter pylori Urease / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.1 MDa |
-Macromolecule #1: Urease subunit alpha
| Macromolecule | Name: Urease subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO / EC number: urease |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.645703 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKLTPKELDK LMLHYAGELA RKRKEKGIKL NYVEAVALIS AHIMEEARAG KKTAAELMQE GRTLLKPDDV MDGVASMIHE VGIEAMFPD GTKLVTVHTP IEANGKLVPG ELFLKNEDIT INEGKKAVSV KVKNVGDRPV QIGSHFHFFE VNRCLDFDRE K TFGKRLDI ...String: MKLTPKELDK LMLHYAGELA RKRKEKGIKL NYVEAVALIS AHIMEEARAG KKTAAELMQE GRTLLKPDDV MDGVASMIHE VGIEAMFPD GTKLVTVHTP IEANGKLVPG ELFLKNEDIT INEGKKAVSV KVKNVGDRPV QIGSHFHFFE VNRCLDFDRE K TFGKRLDI ASGTAVRFEP GEEKSVELID IGGNRRIFGF NALVDRQADN ESKKIALHRA KERGFHGTKS DDNYVKTIKE UniProtKB: Urease subunit alpha |
-Macromolecule #2: Urease subunit beta
| Macromolecule | Name: Urease subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 12 / Enantiomer: LEVO / EC number: urease |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 61.832531 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKKISRKEYV SMYGPTTGDK VRLGDTDLIA EVEHDYTIYG EELKFGGGKT LREGMSQSNN PSKEELDLII TNALIVDYTG IYKADIGIK DGKIAGIGKG GNKDMQDGVK NNLSVGPATE ALAGEGLIVT AGGIDTHIHF ISPQQIPTAF ASGVTTMIGG G TGPADGTN ...String: MKKISRKEYV SMYGPTTGDK VRLGDTDLIA EVEHDYTIYG EELKFGGGKT LREGMSQSNN PSKEELDLII TNALIVDYTG IYKADIGIK DGKIAGIGKG GNKDMQDGVK NNLSVGPATE ALAGEGLIVT AGGIDTHIHF ISPQQIPTAF ASGVTTMIGG G TGPADGTN ATTITPGRRN LKWMLRAAEE YSMNLGFLAK GNTSNDASLA DQIEAGAIGF (KCX)IHEDWGTTP SAINHALD V ADKYDVQVAI HTDTLNEAGC VEDTMAAIAG RTMHTFHTEG AGGGHAPDII KVAGEHNILP ASTNPTIPFT VNTEAEHMD MLMVCHHLDK SIKEDVQFAD SRIRPQTIAA EDTLHDMGIF SITSSDSQAM GRVGEVITRT WQTADKNKKE FGRLKEEKGD NDNFRIKRY LSKYTINPAI AHGISEYVGS VEVGKVADLV LWSPAFFGVK PNMIIKGGFI ALSQMGDANA SIPTPQPVYY R EMFAHHGK AKYDANITFV SQAAYDKGIK EELGLERQVL PVKNCRNITK KDMQFNDTTA HIEVNPETYH VFVDGKEVTS KP ANKVSLA QLFSIF UniProtKB: Urease subunit beta |
-Macromolecule #3: NICKEL (II) ION
| Macromolecule | Name: NICKEL (II) ION / type: ligand / ID: 3 / Number of copies: 24 / Formula: NI |
|---|---|
| Molecular weight | Theoretical: 58.693 Da |
| Chemical component information |  ChemComp-NI: |
-Macromolecule #4: 2-{[1-(3,5-dimethylphenyl)-1H-imidazol-2-yl]sulfanyl}-N-hydroxyac...
| Macromolecule | Name: 2-{[1-(3,5-dimethylphenyl)-1H-imidazol-2-yl]sulfanyl}-N-hydroxyacetamide type: ligand / ID: 4 / Number of copies: 12 / Formula: DJM |
|---|---|
| Molecular weight | Theoretical: 277.342 Da |
| Chemical component information | 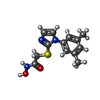 ChemComp-DJM: |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 3191 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 / Component:
| ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 70 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number grids imaged: 1 / Number real images: 956 / Average electron dose: 50.0 e/Å2 / Details: Used 718 movies |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT | ||||||
| Output model |  PDB-6zja: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)