+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12236 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Reprocessing of EMPIAR-10389 data (urease) with Scipion | |||||||||
 Map data Map data | Map produced by Relion plugin in Scipion, filtered according to local resolution. The hand was flipped and the map was realigned to match EMD-10835 | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationurease complex / urease / urease activity / urea catabolic process / nickel cation binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Yersinia enterocolitica (bacteria) Yersinia enterocolitica (bacteria) | |||||||||
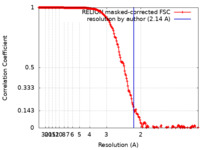
| Method | single particle reconstruction / cryo EM / Resolution: 2.14 Å | |||||||||
 Authors Authors | Sharov G / Morado DR / Carroni M / de la Rosa-Trevin JM | |||||||||
 Citation Citation |  Journal: Acta Crystallogr D Struct Biol / Year: 2021 Journal: Acta Crystallogr D Struct Biol / Year: 2021Title: Using RELION software within the Scipion framework. Authors: Grigory Sharov / Dustin R Morado / Marta Carroni / José Miguel de la Rosa-Trevín /   Abstract: Scipion is a modular image-processing framework that integrates several software packages under a unified interface while taking care of file formats and conversions. Here, new developments and ...Scipion is a modular image-processing framework that integrates several software packages under a unified interface while taking care of file formats and conversions. Here, new developments and capabilities of the Scipion plugin for the widely used RELION software package are presented and illustrated with an image-processing pipeline for published data. The user interfaces of Scipion and RELION are compared and the key differences are highlighted, allowing this manuscript to be used as a guide for both new and experienced users of this software. Different on-the-fly image-processing options are also discussed, demonstrating the flexibility of the Scipion framework. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12236.map.gz emd_12236.map.gz | 309 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12236-v30.xml emd-12236-v30.xml emd-12236.xml emd-12236.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12236_fsc.xml emd_12236_fsc.xml | 18.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_12236.png emd_12236.png | 280.9 KB | ||
| Masks |  emd_12236_msk_1.map emd_12236_msk_1.map | 512 MB |  Mask map Mask map | |
| Others |  emd_12236_additional_1.map.gz emd_12236_additional_1.map.gz emd_12236_half_map_1.map.gz emd_12236_half_map_1.map.gz emd_12236_half_map_2.map.gz emd_12236_half_map_2.map.gz | 266.9 MB 411.9 MB 411.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12236 http://ftp.pdbj.org/pub/emdb/structures/EMD-12236 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12236 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12236 | HTTPS FTP |
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12236.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12236.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map produced by Relion plugin in Scipion, filtered according to local resolution. The hand was flipped and the map was realigned to match EMD-10835 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.639 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12236_msk_1.map emd_12236_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Map produced by Relion plugin in Scipion, postprocessed.
| File | emd_12236_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map produced by Relion plugin in Scipion, postprocessed. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 1 for postprocessing, produced by Relion plugin in Scipion
| File | emd_12236_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 1 for postprocessing, produced by Relion plugin in Scipion | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map 2 for postprocessing, produced by Relion plugin in Scipion
| File | emd_12236_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map 2 for postprocessing, produced by Relion plugin in Scipion | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Urease oligomer from Y. enterocolitica
| Entire | Name: Urease oligomer from Y. enterocolitica |
|---|---|
| Components |
|
-Supramolecule #1: Urease oligomer from Y. enterocolitica
| Supramolecule | Name: Urease oligomer from Y. enterocolitica / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Yersinia enterocolitica (bacteria) Yersinia enterocolitica (bacteria) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.39 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293.15 K / Instrument: FEI VITROBOT MARK IV / Details: 3 sec blot time. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number real images: 4495 / Average exposure time: 8.0 sec. / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)