+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10832 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of V. alginolyticus PomAB | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Vibrio alginolyticus (bacteria) Vibrio alginolyticus (bacteria) | |||||||||
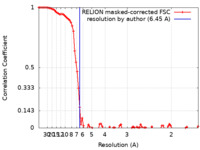
| Method | single particle reconstruction / cryo EM / Resolution: 6.45 Å | |||||||||
 Authors Authors | Santiveri M / Roa-Eguiara A / Taylor NMI | |||||||||
| Funding support |  Denmark, 2 items Denmark, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2020 Journal: Cell / Year: 2020Title: Structure and Function of Stator Units of the Bacterial Flagellar Motor. Authors: Mònica Santiveri / Aritz Roa-Eguiara / Caroline Kühne / Navish Wadhwa / Haidai Hu / Howard C Berg / Marc Erhardt / Nicholas M I Taylor /    Abstract: Many bacteria use the flagellum for locomotion and chemotaxis. Its bidirectional rotation is driven by a membrane-embedded motor, which uses energy from the transmembrane ion gradient to generate ...Many bacteria use the flagellum for locomotion and chemotaxis. Its bidirectional rotation is driven by a membrane-embedded motor, which uses energy from the transmembrane ion gradient to generate torque at the interface between stator units and rotor. The structural organization of the stator unit (MotAB), its conformational changes upon ion transport, and how these changes power rotation of the flagellum remain unknown. Here, we present ~3 Å-resolution cryoelectron microscopy reconstructions of the stator unit in different functional states. We show that the stator unit consists of a dimer of MotB surrounded by a pentamer of MotA. Combining structural data with mutagenesis and functional studies, we identify key residues involved in torque generation and present a detailed mechanistic model for motor function and switching of rotational direction. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10832.map.gz emd_10832.map.gz | 5.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10832-v30.xml emd-10832-v30.xml emd-10832.xml emd-10832.xml | 14.9 KB 14.9 KB | Display Display |  EMDB header EMDB header |
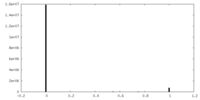
| FSC (resolution estimation) |  emd_10832_fsc.xml emd_10832_fsc.xml | 9.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_10832.png emd_10832.png | 92.1 KB | ||
| Masks |  emd_10832_msk_1.map emd_10832_msk_1.map | 64 MB |  Mask map Mask map | |
| Others |  emd_10832_half_map_1.map.gz emd_10832_half_map_1.map.gz emd_10832_half_map_2.map.gz emd_10832_half_map_2.map.gz | 49.6 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10832 http://ftp.pdbj.org/pub/emdb/structures/EMD-10832 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10832 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10832 | HTTPS FTP |
-Validation report
| Summary document |  emd_10832_validation.pdf.gz emd_10832_validation.pdf.gz | 392.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10832_full_validation.pdf.gz emd_10832_full_validation.pdf.gz | 391.4 KB | Display | |
| Data in XML |  emd_10832_validation.xml.gz emd_10832_validation.xml.gz | 15.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10832 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10832 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10832 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10832 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10832.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10832.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
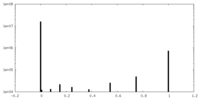
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10832_msk_1.map emd_10832_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
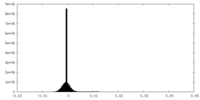
| Density Histograms |
-Half map: #2
| File | emd_10832_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
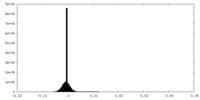
| Density Histograms |
-Half map: #1
| File | emd_10832_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Stator unit PomAB
| Entire | Name: Stator unit PomAB |
|---|---|
| Components |
|
-Supramolecule #1: Stator unit PomAB
| Supramolecule | Name: Stator unit PomAB / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: The stator unit consists of a dimer of PomB surrounded by a pentamer of PomA |
|---|---|
| Source (natural) | Organism:  Vibrio alginolyticus (bacteria) Vibrio alginolyticus (bacteria) |
| Recombinant expression | Organism:  |
-Macromolecule #1: PomA
| Macromolecule | Name: PomA / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio alginolyticus (bacteria) Vibrio alginolyticus (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MDLATLLGLI GGFAFVIMAM VLGGSIGMFV DVTSILIVVG GSIFVVLMKF TMGQFFGATK IAGKAFMFKA DEPEDLIAKI VEMADAARKG GFLALEEMEI NNTFMQKGID LLVDGHDADV VRAALKKDIA LTDERHTQGT GVFRAFGDVA PAMGMIGTLV GLVAMLSNMD ...String: MDLATLLGLI GGFAFVIMAM VLGGSIGMFV DVTSILIVVG GSIFVVLMKF TMGQFFGATK IAGKAFMFKA DEPEDLIAKI VEMADAARKG GFLALEEMEI NNTFMQKGID LLVDGHDADV VRAALKKDIA LTDERHTQGT GVFRAFGDVA PAMGMIGTLV GLVAMLSNMD DPKAIGPAMA VALLTTLYGA ILSNMVFFPI ADKLSLRRDQ ETLNRRLIMD GVLAIQDGQN PRVIDSYLKN YLNEGKRALE IDE |
-Macromolecule #2: PomB
| Macromolecule | Name: PomB / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio alginolyticus (bacteria) Vibrio alginolyticus (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MDDEDNKCDC PPPGLPLWMG TFADLMSLLM CFFVLLLSFS EMDVLKFKQI AGSMKFAFGV QNQLEVKDIP KGTSIIAQEF RPGRPEPTPI DVIMQQTMDI TQQTLEFHEG ESERAGGTKR DEGKLTGGQS PETSTQNNES AEADMQQQQS KEMSQEMETL MESIKKALER ...String: MDDEDNKCDC PPPGLPLWMG TFADLMSLLM CFFVLLLSFS EMDVLKFKQI AGSMKFAFGV QNQLEVKDIP KGTSIIAQEF RPGRPEPTPI DVIMQQTMDI TQQTLEFHEG ESERAGGTKR DEGKLTGGQS PETSTQNNES AEADMQQQQS KEMSQEMETL MESIKKALER EIEQGAIEVE NLGQQIVIRM REKGAFPEGS AFLQPKFRPL VRQIAELVKD VPGIVRVSGH TDNRPLDSEL YRSNWDLSSQ RAVSVAQEME KVRGFSHQRL RVRGMADTEP LLPNDSDENR ALNRRVEISI MQGEPLYSEE VPVIQGTLEV LFQGPGGSGS AWSHPQFEKG GGSGGGSGGS AWSHPQFEK |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 42.82 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)