+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10830 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a protonation mimic of unplugged C. jejuni MotAB | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bacterial flagellar motor / stator unit / locomotion / proton transport / ion transport / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationbacterial-type flagellum-dependent swarming motility / chemotaxis / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Campylobacter jejuni subsp. jejuni 81-176 (Campylobacter) / Campylobacter jejuni subsp. jejuni 81-176 (Campylobacter) /  Campylobacter jejuni subsp. jejuni serotype O:23/36 (strain 81-176) (Campylobacter) Campylobacter jejuni subsp. jejuni serotype O:23/36 (strain 81-176) (Campylobacter) | |||||||||
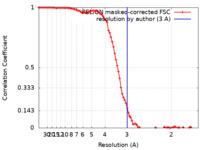
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Santiveri M / Roa-Eguiara A | |||||||||
| Funding support |  Denmark, 2 items Denmark, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2020 Journal: Cell / Year: 2020Title: Structure and Function of Stator Units of the Bacterial Flagellar Motor. Authors: Mònica Santiveri / Aritz Roa-Eguiara / Caroline Kühne / Navish Wadhwa / Haidai Hu / Howard C Berg / Marc Erhardt / Nicholas M I Taylor /    Abstract: Many bacteria use the flagellum for locomotion and chemotaxis. Its bidirectional rotation is driven by a membrane-embedded motor, which uses energy from the transmembrane ion gradient to generate ...Many bacteria use the flagellum for locomotion and chemotaxis. Its bidirectional rotation is driven by a membrane-embedded motor, which uses energy from the transmembrane ion gradient to generate torque at the interface between stator units and rotor. The structural organization of the stator unit (MotAB), its conformational changes upon ion transport, and how these changes power rotation of the flagellum remain unknown. Here, we present ~3 Å-resolution cryoelectron microscopy reconstructions of the stator unit in different functional states. We show that the stator unit consists of a dimer of MotB surrounded by a pentamer of MotA. Combining structural data with mutagenesis and functional studies, we identify key residues involved in torque generation and present a detailed mechanistic model for motor function and switching of rotational direction. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10830.map.gz emd_10830.map.gz | 6.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10830-v30.xml emd-10830-v30.xml emd-10830.xml emd-10830.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10830_fsc.xml emd_10830_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_10830.png emd_10830.png | 108 KB | ||
| Masks |  emd_10830_msk_1.map emd_10830_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10830.cif.gz emd-10830.cif.gz | 5.8 KB | ||
| Others |  emd_10830_half_map_1.map.gz emd_10830_half_map_1.map.gz emd_10830_half_map_2.map.gz emd_10830_half_map_2.map.gz | 49.5 MB 49.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10830 http://ftp.pdbj.org/pub/emdb/structures/EMD-10830 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10830 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10830 | HTTPS FTP |
-Related structure data
| Related structure data |  6ykrMC  6ykmC  6ykpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10830.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10830.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
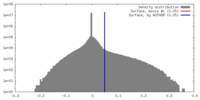
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10830_msk_1.map emd_10830_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_10830_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_10830_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Stator unit MotAB(Delta41-60, D22N)
| Entire | Name: Stator unit MotAB(Delta41-60, D22N) |
|---|---|
| Components |
|
-Supramolecule #1: Stator unit MotAB(Delta41-60, D22N)
| Supramolecule | Name: Stator unit MotAB(Delta41-60, D22N) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: The stator unit consists of a dimer of MotB surrounded by a pentamer of MotA. Single mutation D22N simulates the protonated state of the channel. This stator unit is unplugged (deletion of ...Details: The stator unit consists of a dimer of MotB surrounded by a pentamer of MotA. Single mutation D22N simulates the protonated state of the channel. This stator unit is unplugged (deletion of aminoacids 41 to 60 of MotB). |
|---|---|
| Source (natural) | Organism:  Campylobacter jejuni subsp. jejuni 81-176 (Campylobacter) Campylobacter jejuni subsp. jejuni 81-176 (Campylobacter) |
-Macromolecule #1: Chemotaxis protein MotA, putative
| Macromolecule | Name: Chemotaxis protein MotA, putative / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Campylobacter jejuni subsp. jejuni serotype O:23/36 (strain 81-176) (Campylobacter) Campylobacter jejuni subsp. jejuni serotype O:23/36 (strain 81-176) (Campylobacter)Strain: 81-176 |
| Molecular weight | Theoretical: 28.195816 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDLSTILGMV LAVTSISVGD ILEGGNPLHV IHLSSFLIVM PTAAFCAMTS THKKIVKAAY KELKVVFKGS GVNLPERIAQ LIEFAIIAR RDGLLALESR TNEIENEFLK NAMMMLVDGK SFEEIHESME IQTEQLEEHY KECAEYWIVF GETCPTMGLV G AVFGLILA ...String: MDLSTILGMV LAVTSISVGD ILEGGNPLHV IHLSSFLIVM PTAAFCAMTS THKKIVKAAY KELKVVFKGS GVNLPERIAQ LIEFAIIAR RDGLLALESR TNEIENEFLK NAMMMLVDGK SFEEIHESME IQTEQLEEHY KECAEYWIVF GETCPTMGLV G AVFGLILA LKLLDNPQAM AAGISGAFTA TVTGIFGAYA LFAPWGKKLK ANGMDLVKEQ IVITEAIKGI AEGANPRDLE AK LFNFLSH DDPRISQFDK G UniProtKB: Chemotaxis protein MotA, putative |
-Macromolecule #2: Chemotaxis protein MotB, putative
| Macromolecule | Name: Chemotaxis protein MotB, putative / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Campylobacter jejuni subsp. jejuni serotype O:23/36 (strain 81-176) (Campylobacter) Campylobacter jejuni subsp. jejuni serotype O:23/36 (strain 81-176) (Campylobacter)Strain: 81-176 |
| Molecular weight | Theoretical: 29.862643 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAKKHKCPEC PAGEKWAVPY ANFLSLLLAL FIALWAISKT TQTVKEESKT QEKYKGAAKE ESDELKSLKQ MTMTQQETIK RLQAALDQS DNQVALNLPS KVEFERGSAQ IVSADIQDYL KRMAELTTYL PPQAKIEIRG YTDNSDSIIR SYELAYQRAE N VLKYFIEG ...String: MAKKHKCPEC PAGEKWAVPY ANFLSLLLAL FIALWAISKT TQTVKEESKT QEKYKGAAKE ESDELKSLKQ MTMTQQETIK RLQAALDQS DNQVALNLPS KVEFERGSAQ IVSADIQDYL KRMAELTTYL PPQAKIEIRG YTDNSDSIIR SYELAYQRAE N VLKYFIEG GANLKNISIK SYGLNNPING NPQALENNRV EIYFKVDTAD TSTQKSVLEL INKIGTKAPG TLEVLFQGPG GS GSAWSHP QFEKGGGSGG GSGGSAWSHP QFEK UniProtKB: Chemotaxis protein MotB, putative |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 6 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.68 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R2/1 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 43.44 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)