[English] 日本語
 Yorodumi
Yorodumi- EMDB-10810: In-situ structure of the trimeric HEF from influenza C viral part... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10810 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | In-situ structure of the trimeric HEF from influenza C viral particles with a matrix layer by cryo-ET | ||||||||||||
 Map data Map data | Filtered map | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Receptor / esterase / fusion / influenza / VIRAL PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationsialate 9-O-acetylesterase activity / sialate 4-O-acetylesterase activity / sialate O-acetylesterase / viral budding from plasma membrane / host cell surface receptor binding / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell ...sialate 9-O-acetylesterase activity / sialate 4-O-acetylesterase activity / sialate O-acetylesterase / viral budding from plasma membrane / host cell surface receptor binding / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / membrane Similarity search - Function | ||||||||||||
| Biological species |  Influenza C virus (strain C/Johannesburg/1/1966) / Influenza C virus (strain C/Johannesburg/1/1966) /  Influenza C virus Influenza C virus | ||||||||||||
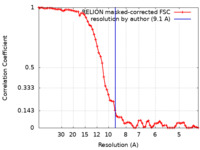
| Method | subtomogram averaging / cryo EM / Resolution: 9.1 Å | ||||||||||||
 Authors Authors | Halldorsson S / Rosenthal PB | ||||||||||||
| Funding support |  United Kingdom, 3 items United Kingdom, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: In situ structure and organization of the influenza C virus surface glycoprotein. Authors: Steinar Halldorsson / Kasim Sader / Jack Turner / Lesley J Calder / Peter B Rosenthal /   Abstract: The lipid-enveloped influenza C virus contains a single surface glycoprotein, the haemagglutinin-esterase-fusion (HEF) protein, that mediates receptor binding, receptor destruction, and membrane ...The lipid-enveloped influenza C virus contains a single surface glycoprotein, the haemagglutinin-esterase-fusion (HEF) protein, that mediates receptor binding, receptor destruction, and membrane fusion at the low pH of the endosome. Here we apply electron cryotomography and subtomogram averaging to describe the structural basis for hexagonal lattice formation by HEF on the viral surface. The conformation of the glycoprotein in situ is distinct from the structure of the isolated trimeric ectodomain, showing that a splaying of the membrane distal domains is required to mediate contacts that form the lattice. The splaying of these domains is also coupled to changes in the structure of the stem region which is involved in membrane fusion, thereby linking HEF's membrane fusion conformation with its assembly on the virus surface. The glycoprotein lattice can form independent of other virion components but we show a major role for the matrix layer in particle formation. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10810.map.gz emd_10810.map.gz | 28.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10810-v30.xml emd-10810-v30.xml emd-10810.xml emd-10810.xml | 22.1 KB 22.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10810_fsc.xml emd_10810_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_10810.png emd_10810.png | 96.5 KB | ||
| Masks |  emd_10810_msk_1.map emd_10810_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10810.cif.gz emd-10810.cif.gz | 6.5 KB | ||
| Others |  emd_10810_additional_1.map.gz emd_10810_additional_1.map.gz emd_10810_half_map_1.map.gz emd_10810_half_map_1.map.gz emd_10810_half_map_2.map.gz emd_10810_half_map_2.map.gz | 23.2 MB 23.3 MB 23.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10810 http://ftp.pdbj.org/pub/emdb/structures/EMD-10810 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10810 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10810 | HTTPS FTP |
-Validation report
| Summary document |  emd_10810_validation.pdf.gz emd_10810_validation.pdf.gz | 803.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10810_full_validation.pdf.gz emd_10810_full_validation.pdf.gz | 803 KB | Display | |
| Data in XML |  emd_10810_validation.xml.gz emd_10810_validation.xml.gz | 12.7 KB | Display | |
| Data in CIF |  emd_10810_validation.cif.gz emd_10810_validation.cif.gz | 17.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10810 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10810 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10810 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10810 | HTTPS FTP |
-Related structure data
| Related structure data |  6yi5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10810.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10810.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Filtered map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10810_msk_1.map emd_10810_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unfiltered map
| File | emd_10810_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_10810_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_10810_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Influenza C virus
| Entire | Name:  Influenza C virus Influenza C virus |
|---|---|
| Components |
|
-Supramolecule #1: Influenza C virus
| Supramolecule | Name: Influenza C virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Purified from infected cells. / NCBI-ID: 100673 / Sci species name: Influenza C virus / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Hemagglutinin-esterase-fusion glycoprotein
| Macromolecule | Name: Hemagglutinin-esterase-fusion glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO / EC number: sialate O-acetylesterase |
|---|---|
| Source (natural) | Organism:  Influenza C virus (strain C/Johannesburg/1/1966) Influenza C virus (strain C/Johannesburg/1/1966) |
| Molecular weight | Theoretical: 48.230699 KDa |
| Sequence | String: EKIKICLQKQ VNSSFSLHNG FGGNLYATEE KRMFELVKPK AGASVLNQST WIGFGDSRTD KSNSAFPRSA DVSAKTADKF RFLSGGSLM LSMFGPPGKV DYLYQGCGKH KVFYEGVNWS PHAAINCYRK NWTDIKLNFQ KNIYELASQS HCMSLVNALD K TIPLQVTA ...String: EKIKICLQKQ VNSSFSLHNG FGGNLYATEE KRMFELVKPK AGASVLNQST WIGFGDSRTD KSNSAFPRSA DVSAKTADKF RFLSGGSLM LSMFGPPGKV DYLYQGCGKH KVFYEGVNWS PHAAINCYRK NWTDIKLNFQ KNIYELASQS HCMSLVNALD K TIPLQVTA GTAGNCNNSF LKNPALYTQE VKPSENKCGK ENLAFFTLPT QFGTYECKLH LVASCYFIYD SKEVYNKRGC DN YFQVIYD SFGKVVGGLD NRVSPYTGNS GDTPTMQCDM LQLKPGRYSV RSSPRFLLMP ERSYCFDMKE KGPVTAVQSI WGK GRESDY AVDQACLSTP GCMLIQKQKP YIGEADDHHG DQEMRELLSG LDYEARCISQ SGWVNETSPF TEKYLLPPKF GRCP LAAKE ESIPKIPDGL LIPTSGTDTT VTKPKSR UniProtKB: Hemagglutinin-esterase-fusion glycoprotein |
-Macromolecule #2: Hemagglutinin-esterase-fusion glycoprotein
| Macromolecule | Name: Hemagglutinin-esterase-fusion glycoprotein / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO / EC number: sialate O-acetylesterase |
|---|---|
| Source (natural) | Organism:  Influenza C virus (strain C/Johannesburg/1/1966) Influenza C virus (strain C/Johannesburg/1/1966) |
| Molecular weight | Theoretical: 18.977541 KDa |
| Sequence | String: IFGIDDLIIG VLFVAIVETG IGGYLLGSRK ESGGGVTKES AEKGFEKIGN DIQILKSSIN IAIEKLNDRI SHDEQAIRDL TLEIENARS EALLGELGII RALLVGNISI GLQESLWELA SEITNRAGDL AVEVSPGCWI IDNNICDQSC QNFIFKFNET A PVPTIPPL DTKIDLQ UniProtKB: Hemagglutinin-esterase-fusion glycoprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III | ||||||||||||
| Details | Heterogeneous mix of viral particles, viral like particles and cellular vesicles |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Number grids imaged: 2 / Average exposure time: 1.57 sec. / Average electron dose: 1.57 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 0.0045000000000000005 µm / Nominal defocus min: 0.0025 µm / Nominal magnification: 64000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)