[English] 日本語
 Yorodumi
Yorodumi- EMDB-10795: Negative stain map of S. cerevisiae Brf1 and TBP bound to the TFI... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10795 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative stain map of S. cerevisiae Brf1 and TBP bound to the TFIIIC-subcomplex tauA | |||||||||
 Map data Map data | Refined map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 32.36 Å | |||||||||
 Authors Authors | Vorlaender MK / Muller CW | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Structure of the TFIIIC subcomplex τA provides insights into RNA polymerase III pre-initiation complex formation. Authors: Matthias K Vorländer / Anna Jungblut / Kai Karius / Florence Baudin / Helga Grötsch / Jan Kosinski / Christoph W Müller /  Abstract: Transcription factor (TF) IIIC is a conserved eukaryotic six-subunit protein complex with dual function. It serves as a general TF for most RNA polymerase (Pol) III genes by recruiting TFIIIB, but it ...Transcription factor (TF) IIIC is a conserved eukaryotic six-subunit protein complex with dual function. It serves as a general TF for most RNA polymerase (Pol) III genes by recruiting TFIIIB, but it is also involved in chromatin organization and regulation of Pol II genes through interaction with CTCF and condensin II. Here, we report the structure of the S. cerevisiae TFIIIC subcomplex τA, which contains the most conserved subunits of TFIIIC and is responsible for recruitment of TFIIIB and transcription start site (TSS) selection at Pol III genes. We show that τA binding to its promoter is auto-inhibited by a disordered acidic tail of subunit τ95. We further provide a negative-stain reconstruction of τA bound to the TFIIIB subunits Brf1 and TBP. This shows that a ruler element in τA achieves positioning of TFIIIB upstream of the TSS, and suggests remodeling of the complex during assembly of TFIIIB by TFIIIC. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10795.map.gz emd_10795.map.gz | 23.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10795-v30.xml emd-10795-v30.xml emd-10795.xml emd-10795.xml | 21.8 KB 21.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10795_fsc.xml emd_10795_fsc.xml | 9.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_10795.png emd_10795.png | 78.7 KB | ||
| Others |  emd_10795_half_map_1.map.gz emd_10795_half_map_1.map.gz emd_10795_half_map_2.map.gz emd_10795_half_map_2.map.gz | 23.4 MB 23.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10795 http://ftp.pdbj.org/pub/emdb/structures/EMD-10795 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10795 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10795 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10795.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10795.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refined map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.292 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
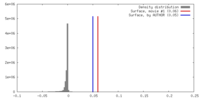
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Half map 2
| File | emd_10795_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
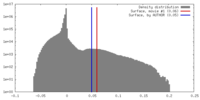
| Density Histograms |
-Half map: Half map 1
| File | emd_10795_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
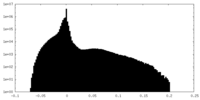
| Density Histograms |
- Sample components
Sample components
-Entire : Complex between Brf1, TBP, and the tauA subcomplex of TFIIIC
| Entire | Name: Complex between Brf1, TBP, and the tauA subcomplex of TFIIIC |
|---|---|
| Components |
|
-Supramolecule #1: Complex between Brf1, TBP, and the tauA subcomplex of TFIIIC
| Supramolecule | Name: Complex between Brf1, TBP, and the tauA subcomplex of TFIIIC type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Crosslinked with Glutaraldehyde in absence of DNA |
|---|---|
| Molecular weight | Theoretical: 300 KDa |
-Supramolecule #2: Transcription factor tau 131 kDa subunit, Transcription factor ta...
| Supramolecule | Name: Transcription factor tau 131 kDa subunit, Transcription factor tau 95 kDa subunit and Transcription factor tau 55 kDa subunit type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
-Supramolecule #3: Transcription factor IIIB 70 kDa subunit and TATA-box-binding protein
| Supramolecule | Name: Transcription factor IIIB 70 kDa subunit and TATA-box-binding protein type: complex / ID: 3 / Parent: 1 / Macromolecule list: #4-#5 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Macromolecule #1: Transcription factor tau 131 kDa subunit
| Macromolecule | Name: Transcription factor tau 131 kDa subunit / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAAGKLKKEQ QNQSAERESA DTGKVNDEDE EHLYGNIDDY KHLIQDEEYD DEDVPHDLQL SEDEYNSER DSSLLAEFSD YGEISEDDEE DFMNAIREAS NFKVKKKKKN DKGKSYGRQR K ERVLDPEV AQLLSQANEA FVRNDLQVAE RLFNEVIKKD ARNFAAYETL ...String: MAAGKLKKEQ QNQSAERESA DTGKVNDEDE EHLYGNIDDY KHLIQDEEYD DEDVPHDLQL SEDEYNSER DSSLLAEFSD YGEISEDDEE DFMNAIREAS NFKVKKKKKN DKGKSYGRQR K ERVLDPEV AQLLSQANEA FVRNDLQVAE RLFNEVIKKD ARNFAAYETL GDIYQLQGRL ND CCNSWFL AAHLNASDWE FWKIVAILSA DLDHVRQAIY CFSRVISLNP MEWESIYRRS MLY KKTGQL ARALDGFQRL YMYNPYDANI LRELAILYVD YDRIEDSIEL YMKVFNANVE RREA ILAAL ENALDSSDEE SAAEGEDADE KEPLEQDEDR QMFPDINWKK IDAKYKCIPF DWSSL NILA ELFLKLAVSE VDGIKTIKKC ARWIQRRESQ TFWDHVPDDS EFDNRRFKNS TFDSLL AAE KEKSYNIPID IRVRLGLLRL NTDNLVEALN HFQCLYDETF SDVADLYFEA ATALTRA EK YKEAIDFFTP LLSLEEWRTT DVFKPLARCY KEIESYETAK EFYELAIKSE PDDLDIRV S LAEVYYRLND PETFKHMLVD VVEMRKHQVD ETLHRISNEK SSNDTSDISS KPLLEDSKF RTFRKKKRTP YDAERERIER ERRITAKVVD KYEKMKKFEL NSGLNEAKQA SIWINTVSEL VDIFSSVKN FFMKSRSRKF VGILRRTKKF NTELDFQIER LSKLAEGDSV FEGPLMEERV T LTSATELR GLSYEQWFEL FMELSLVIAK YQSVEDGLSV VETAQEVNVF FQDPERVKMM KF VKLAIVL QMDDEEELAE NLRGLLNQFQ FNRKVLQVFM YSLCRGPSSL NILSSTIQQK FFL RQLKAF DSCRYNTEVN GQASITNKEV YNPNKKSSPY LYYIYAVLLY SSRGFLSALQ YLTR LEEDI PDDPMVNLLM GLSHIHRAMQ RLTAQRHFQI FHGLRYLYRY HKIRKSLYTD LEKQE ADYN LGRAFHLIGL VSIAIEYYNR VLENYDDGKL KKHAAYNSII IYQQSGNVEL ADHLME KYL SIRSGG |
-Macromolecule #2: Transcription factor tau 95 kDa subunit
| Macromolecule | Name: Transcription factor tau 95 kDa subunit / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MHHHHHHENL YFQSMPVEEP LATLSSIPDS SADQAPPLIA DEFTLDLPRI PSLELPLNVS TKHSSIQKA IKMCGGIEKV KEAFKEHGPI ESQHGLQLYL NDDTDSDGSK SYFNEHPVIG K RVPFRDES VILKVTMPKG TLSKNNNSVK DSIKSLKDSN KLRVTPVSIV ...String: MHHHHHHENL YFQSMPVEEP LATLSSIPDS SADQAPPLIA DEFTLDLPRI PSLELPLNVS TKHSSIQKA IKMCGGIEKV KEAFKEHGPI ESQHGLQLYL NDDTDSDGSK SYFNEHPVIG K RVPFRDES VILKVTMPKG TLSKNNNSVK DSIKSLKDSN KLRVTPVSIV DNTIKFREMS DF QIKLDNV PSAREFKSSF GSLEWNNFKS FVNSVPDNDS QPQENIGNLI LDRSVKIPST DFQ LPPPPK LSMVGFPLLY KYKANPFAKK KKNGVTEVKG TYIKNYQLFV HDLSDKTVIP SQAH EQVLY DFEVAKKTKV YPGTKSDSKF YESLEECLKI LRELFARRPI WVKRHLDGIV PKKIH HTMK IALALISYRF TMGPWRNTYI KFGIDPRSSV EYAQYQTEYF KIERKLLSSP IVKKNV PKP PPLVFESDTP GGIDSRFKFD GKRIPWYLML QIDLLIGEPN IAEVFHNVEY LDKANEL TG WFKELDLVKI RRIVKYELGC MVQGNYEYNK YKLKYFKTML FVKESMVPEN KNSEEGMG V NTNKDADGDI NMDAGSQMSS NAIEEDKGIA AGDDFDDNGA ITEEPDDAAL ENEEMDTDQ NLKVPASIDD DVDDVDADEE EQESFDVKTA SFQDIINKIA KLDPKTAETM KSELKGFVDE VDL |
-Macromolecule #3: Transcription factor tau 55 kDa subunit
| Macromolecule | Name: Transcription factor tau 55 kDa subunit / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MVVNTIYIAR HGYRSNWLPE GPYPDPLTGI DSDVPLAEHG VQQAKELAHY LLSLDNQPEA AFASPFYRC LETVQPIAKL LEIPVYLERG IGEWYRPDRK PVIPVPAGYE ILSKFFPGVI S QEWDSTLT PNEKGETEQE MYMRFKKFWP LFIERVEKEY PNVECILLVT ...String: MVVNTIYIAR HGYRSNWLPE GPYPDPLTGI DSDVPLAEHG VQQAKELAHY LLSLDNQPEA AFASPFYRC LETVQPIAKL LEIPVYLERG IGEWYRPDRK PVIPVPAGYE ILSKFFPGVI S QEWDSTLT PNEKGETEQE MYMRFKKFWP LFIERVEKEY PNVECILLVT HAASKIALGM SL LGYDNPR MSLNENGDKI RSGSCSLDKY EILKKSYDTI DETDDQTSFT YIPFSDRKWV LTM NGNTEF LSSGEEMNWN FDCVAEAGSD ADIKKRQMTK KTSSPIPEAD DQTEVETVYI SVDI PSGNY KERTEIAKSA ILQYSGLETD APLFRIGNRL YEGSWERLVG TELAFPNAAH VHKKT AGLL SPTEENETTN AGQSKGSSTA NDPNIQIQEE DVGLPDSTNT SRDHTGDKEE VQSEKI YRI KERIVLSNVR PM |
-Macromolecule #4: Transcription factor IIIB 70 kDa subunit
| Macromolecule | Name: Transcription factor IIIB 70 kDa subunit / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MSALESREAT LNNARRKLRA VSYALHIPEY ITDAAFQWYK LALANNFVQG R RSQNVIAS CLYVACRKEK THHMLIDFSS RLQVSVYSIG ATFLKMVKKL HITELPLADP SL FIQHFAE KLDLADKKIK VVKDAVKLAQ RMSKDWMFEG RRPAGIAGAC ILLACRMNNL RRT ...String: MSALESREAT LNNARRKLRA VSYALHIPEY ITDAAFQWYK LALANNFVQG R RSQNVIAS CLYVACRKEK THHMLIDFSS RLQVSVYSIG ATFLKMVKKL HITELPLADP SL FIQHFAE KLDLADKKIK VVKDAVKLAQ RMSKDWMFEG RRPAGIAGAC ILLACRMNNL RRT HTEIVA VSHVAEETLQ QRLNEFKNTK AAKLSVQKFR ENDVEDGEAR PPSFVKNRKK ERKI KDSLD KEEMFQTSEE ALNKNPILTQ VLGEQELSSK EVLFYLKQFS ERRARVVERI KATNG IDGE NIYHEGSENE TRKRKLSEVS IQNEHVEGED KETEGTEEKV KKVKTKTSEE KKENES GHF QDAIDGYSLE TDPYCPRNLH LLPTTDTYLS KVSDDPDNLE DVDDEELNAH LLNEEAS KL KERIWIGLNA DFLLEQESKR LKQEADIATG NTSVKKKRTR RRNNTRSDEP TKTVDAAA A IGLMSDLQDK SGLHAALKAA EESGDFTTAD SVKNMLQKAS FSKKINYDAI DGLFRLEHHH HHH |
-Macromolecule #5: TATA-box-binding protein
| Macromolecule | Name: TATA-box-binding protein / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: SGIVPTLQNI VATVTLGCRL DLKTVALHAR NAEYNPKRFA AVIMRIREPK TTALIFASGK MVVTGAKSE DDSKLASRKY ARIIQKIGFA AKFTDFKIQN IVGSCDVKFP IRLEGLAFSH G TFSSYEPE LFPGLIYRMV KPKIVLLIFV SGKIVLTGAK QREEIYQAFE AIYPVLSEFR KM |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl Acetate |
| Details | Crosslinked sample |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT / Target criteria: Envelope score |
|---|
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)