+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10817 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the TFIIIC subcomplex tauA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TFIIIC / tauA / Transcription initiation / Pol III / TFIIIB / Transcription factor / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology information5S class rRNA transcription by RNA polymerase III / transcription factor TFIIIC complex / RNA Polymerase III Transcription Initiation From Type 2 Promoter / transcription initiation at RNA polymerase III promoter / phosphatase activity / transcription by RNA polymerase III / DNA binding / nucleoplasm / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
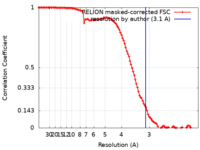
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Vorlaender MK / Muller CW | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Structure of the TFIIIC subcomplex τA provides insights into RNA polymerase III pre-initiation complex formation. Authors: Matthias K Vorländer / Anna Jungblut / Kai Karius / Florence Baudin / Helga Grötsch / Jan Kosinski / Christoph W Müller /  Abstract: Transcription factor (TF) IIIC is a conserved eukaryotic six-subunit protein complex with dual function. It serves as a general TF for most RNA polymerase (Pol) III genes by recruiting TFIIIB, but it ...Transcription factor (TF) IIIC is a conserved eukaryotic six-subunit protein complex with dual function. It serves as a general TF for most RNA polymerase (Pol) III genes by recruiting TFIIIB, but it is also involved in chromatin organization and regulation of Pol II genes through interaction with CTCF and condensin II. Here, we report the structure of the S. cerevisiae TFIIIC subcomplex τA, which contains the most conserved subunits of TFIIIC and is responsible for recruitment of TFIIIB and transcription start site (TSS) selection at Pol III genes. We show that τA binding to its promoter is auto-inhibited by a disordered acidic tail of subunit τ95. We further provide a negative-stain reconstruction of τA bound to the TFIIIB subunits Brf1 and TBP. This shows that a ruler element in τA achieves positioning of TFIIIB upstream of the TSS, and suggests remodeling of the complex during assembly of TFIIIB by TFIIIC. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10817.map.gz emd_10817.map.gz | 62.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10817-v30.xml emd-10817-v30.xml emd-10817.xml emd-10817.xml | 20.9 KB 20.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10817_fsc.xml emd_10817_fsc.xml | 10.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_10817.png emd_10817.png | 140.1 KB | ||
| Filedesc metadata |  emd-10817.cif.gz emd-10817.cif.gz | 7.5 KB | ||
| Others |  emd_10817_half_map_1.map.gz emd_10817_half_map_1.map.gz emd_10817_half_map_2.map.gz emd_10817_half_map_2.map.gz | 81.5 MB 81.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10817 http://ftp.pdbj.org/pub/emdb/structures/EMD-10817 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10817 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10817 | HTTPS FTP |
-Related structure data
| Related structure data |  6yj6MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10817.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10817.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.041 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
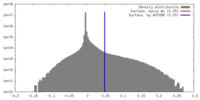
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_10817_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
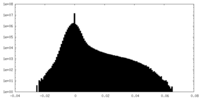
| Density Histograms |
-Half map: #2
| File | emd_10817_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : TFIIIC subcomplex tauA
| Entire | Name: TFIIIC subcomplex tauA |
|---|---|
| Components |
|
-Supramolecule #1: TFIIIC subcomplex tauA
| Supramolecule | Name: TFIIIC subcomplex tauA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Generated by co-expression of subunits in insect cells |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 245 KDa |
-Macromolecule #1: Transcription factor tau 131 kDa subunit
| Macromolecule | Name: Transcription factor tau 131 kDa subunit / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 120.746969 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAAGKLKKEQ QNQSAERESA DTGKVNDEDE EHLYGNIDDY KHLIQDEEYD DEDVPHDLQL SEDEYNSERD SSLLAEFSDY GEISEDDEE DFMNAIREAS NFKVKKKKKN DKGKSYGRQR KERVLDPEVA QLLSQANEAF VRNDLQVAER LFNEVIKKDA R NFAAYETL ...String: MAAGKLKKEQ QNQSAERESA DTGKVNDEDE EHLYGNIDDY KHLIQDEEYD DEDVPHDLQL SEDEYNSERD SSLLAEFSDY GEISEDDEE DFMNAIREAS NFKVKKKKKN DKGKSYGRQR KERVLDPEVA QLLSQANEAF VRNDLQVAER LFNEVIKKDA R NFAAYETL GDIYQLQGRL NDCCNSWFLA AHLNASDWEF WKIVAILSAD LDHVRQAIYC FSRVISLNPM EWESIYRRSM LY KKTGQLA RALDGFQRLY MYNPYDANIL RELAILYVDY DRIEDSIELY MKVFNANVER REAILAALEN ALDSSDEESA AEG EDADEK EPLEQDEDRQ MFPDINWKKI DAKYKCIPFD WSSLNILAEL FLKLAVSEVD GIKTIKKCAR WIQRRESQTF WDHV PDDSE FDNRRFKNST FDSLLAAEKE KSYNIPIDIR VRLGLLRLNT DNLVEALNHF QCLYDETFSD VADLYFEAAT ALTRA EKYK EAIDFFTPLL SLEEWRTTDV FKPLARCYKE IESYETAKEF YELAIKSEPD DLDIRVSLAE VYYRLNDPET FKHMLV DVV EMRKHQVDET LHRISNEKSS NDTSDISSKP LLEDSKFRTF RKKKRTPYDA ERERIERERR ITAKVVDKYE KMKKFEL NS GLNEAKQASI WINTVSELVD IFSSVKNFFM KSRSRKFVGI LRRTKKFNTE LDFQIERLSK LAEGDSVFEG PLMEERVT L TSATELRGLS YEQWFELFME LSLVIAKYQS VEDGLSVVET AQEVNVFFQD PERVKMMKFV KLAIVLQMDD EEELAENLR GLLNQFQFNR KVLQVFMYSL CRGPSSLNIL SSTIQQKFFL RQLKAFDSCR YNTEVNGQAS ITNKEVYNPN KKSSPYLYYI YAVLLYSSR GFLSALQYLT RLEEDIPDDP MVNLLMGLSH IHRAMQRLTA QRHFQIFHGL RYLYRYHKIR KSLYTDLEKQ E ADYNLGRA FHLIGLVSIA IEYYNRVLEN YDDGKLKKHA AYNSIIIYQQ SGNVELADHL MEKYLSIRSG G UniProtKB: Transcription factor tau 131 kDa subunit |
-Macromolecule #2: Transcription factor tau 95 kDa subunit,Transcription factor tau ...
| Macromolecule | Name: Transcription factor tau 95 kDa subunit,Transcription factor tau 95 kDa subunit type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 77.363664 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MHHHHHHENL YFQSMPVEEP LATLSSIPDS SADQAPPLIA DEFTLDLPRI PSLELPLNVS TKHSSIQKAI KMCGGIEKVK EAFKEHGPI ESQHGLQLYL NDDTDSDGSK SYFNEHPVIG KRVPFRDESV ILKVTMPKGT LSKNNNSVKD SIKSLKDSNK L RVTPVSIV ...String: MHHHHHHENL YFQSMPVEEP LATLSSIPDS SADQAPPLIA DEFTLDLPRI PSLELPLNVS TKHSSIQKAI KMCGGIEKVK EAFKEHGPI ESQHGLQLYL NDDTDSDGSK SYFNEHPVIG KRVPFRDESV ILKVTMPKGT LSKNNNSVKD SIKSLKDSNK L RVTPVSIV DNTIKFREMS DFQIKLDNVP SAREFKSSFG SLEWNNFKSF VNSVPDNDSQ PQENIGNLIL DRSVKIPSTD FQ LPPPPKL SMVGFPLLYK YKANPFAKKK KNGVTEVKGT YIKNYQLFVH DLSDKTVIPS QAHEQVLYDF EVAKKTKVYP GTK SDSKFY ESLEECLKIL RELFARRPIW VKRHLDGIVP KKIHHTMKIA LALISYRFTM GPWRNTYIKF GIDPRSSVEY AQYQ TEYFK IERKLLSSPI VKKNVPKPPP LVFESDTPGG IDSRFKFDGK RIPWYLMLQI DLLIGEPNIA EVFHNVEYLD KANEL TGWF KELDLVKIRR IVKYELGCMV QGNYEYNKYK LKYFKTMLFV KESMVPENKN SEEGMGVNTN KDADGDINMD AGSQMS SNA IEEDKGIAAG DDFDDNGAIT EEPDDAALEN EEMDTDQNLK VPASIDDDVD DVDADEEEQE SFDVKTASFQ DIINKIA KL DPKTAETMKS ELKGFVDEVD L(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) UniProtKB: Transcription factor tau 95 kDa subunit |
-Macromolecule #3: Transcription factor tau 55 kDa subunit
| Macromolecule | Name: Transcription factor tau 55 kDa subunit / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 49.198898 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MVVNTIYIAR HGYRSNWLPE GPYPDPLTGI DSDVPLAEHG VQQAKELAHY LLSLDNQPEA AFASPFYRCL ETVQPIAKLL EIPVYLERG IGEWYRPDRK PVIPVPAGYE ILSKFFPGVI SQEWDSTLTP NEKGETEQEM YMRFKKFWPL FIERVEKEYP N VECILLVT ...String: MVVNTIYIAR HGYRSNWLPE GPYPDPLTGI DSDVPLAEHG VQQAKELAHY LLSLDNQPEA AFASPFYRCL ETVQPIAKLL EIPVYLERG IGEWYRPDRK PVIPVPAGYE ILSKFFPGVI SQEWDSTLTP NEKGETEQEM YMRFKKFWPL FIERVEKEYP N VECILLVT HAASKIALGM SLLGYDNPRM SLNENGDKIR SGSCSLDKYE ILKKSYDTID ETDDQTSFTY IPFSDRKWVL TM NGNTEFL SSGEEMNWNF DCVAEAGSDA DIKKRQMTKK TSSPIPEADD QTEVETVYIS VDIPSGNYKE RTEIAKSAIL QYS GLETDA PLFRIGNRLY EGSWERLVGT ELAFPNAAHV HKKTAGLLSP TEENETTNAG QSKGSSTAND PNIQIQEEDV GLPD STNTS RDHTGDKEEV QSEKIYRIKE RIVLSNVRPM UniProtKB: Transcription factor tau 55 kDa subunit |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.3 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 20 mM HEPES, 5 mM DTT, 75 mM NaCl |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 45 sec. / Details: Pelco Easygglow instrument |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV / Details: Blot force 2, Blot time 4. |
| Details | 4 mM CHAPSO added prior to freezing |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-36 / Number grids imaged: 1 / Number real images: 5824 / Average exposure time: 48.73 sec. / Average electron dose: 1.35 e/Å2 / Details: collected 8 frames per hole using beam shift |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -2.0 µm / Nominal defocus min: -0.5 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-6yj6: |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)