+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10675 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 1.9 A reconstruction of mouse heavy chain apoferritin | |||||||||
 Map data Map data | sharpened masked map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationIron uptake and transport / Golgi Associated Vesicle Biogenesis / negative regulation of ferroptosis / ferroxidase / autolysosome / ferroxidase activity / Neutrophil degranulation / endocytic vesicle lumen / ferric iron binding / autophagosome ...Iron uptake and transport / Golgi Associated Vesicle Biogenesis / negative regulation of ferroptosis / ferroxidase / autolysosome / ferroxidase activity / Neutrophil degranulation / endocytic vesicle lumen / ferric iron binding / autophagosome / iron ion transport / intracellular iron ion homeostasis / immune response / iron ion binding / negative regulation of cell population proliferation / mitochondrion / extracellular region / identical protein binding / membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
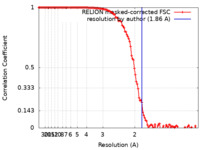
| Method | single particle reconstruction / cryo EM / Resolution: 1.86 Å | |||||||||
 Authors Authors | Fislage M / Shkumatov A / Stroobants A / Efremov R | |||||||||
| Funding support |  Belgium, 2 items Belgium, 2 items
| |||||||||
 Citation Citation |  Journal: IUCrJ / Year: 2020 Journal: IUCrJ / Year: 2020Title: Assessing the JEOL CRYO ARM 300 for high-throughput automated single-particle cryo-EM in a multiuser environment. Authors: Marcus Fislage / Alexander V Shkumatov / Annelore Stroobants / Rouslan G Efremov /  Abstract: Single-particle cryo-EM has become an indispensable structural biology method. It requires regular access to high-resolution electron cryogenic microscopes. To fully utilize the capacity of the ...Single-particle cryo-EM has become an indispensable structural biology method. It requires regular access to high-resolution electron cryogenic microscopes. To fully utilize the capacity of the expensive high-resolution instruments, the time used for data acquisition and the rate of data collection have to be maximized. This in turn requires high stability and high uptime of the instrument. One of the first 300 kV JEOL CRYO ARM 300 microscopes has been installed at the cryo-EM facility BECM at VIB-VUB, Brussels, where the microscope is used for continuous data collection on multiple projects. Here, the suitability and performance of the microscope is assessed for high-throughput single-particle data collection. In particular, the properties of the illumination system, the stage stability and ice contamination rates are reported. The microscope was benchmarked using mouse heavy-chain apoferritin which was reconstructed to a resolution of 1.9 Å. Finally, uptime and throughput statistics of the instrument accumulated during the first six months of the facility operation in user access mode are reported. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10675.map.gz emd_10675.map.gz | 22.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10675-v30.xml emd-10675-v30.xml emd-10675.xml emd-10675.xml | 16.7 KB 16.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10675_fsc.xml emd_10675_fsc.xml | 12 KB | Display |  FSC data file FSC data file |
| Images |  emd_10675.png emd_10675.png | 269.4 KB | ||
| Masks |  emd_10675_msk_1.map emd_10675_msk_1.map | 149.9 MB |  Mask map Mask map | |
| Others |  emd_10675_half_map_1.map.gz emd_10675_half_map_1.map.gz emd_10675_half_map_2.map.gz emd_10675_half_map_2.map.gz | 113.3 MB 113.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10675 http://ftp.pdbj.org/pub/emdb/structures/EMD-10675 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10675 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10675 | HTTPS FTP |
-Validation report
| Summary document |  emd_10675_validation.pdf.gz emd_10675_validation.pdf.gz | 399.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10675_full_validation.pdf.gz emd_10675_full_validation.pdf.gz | 398.2 KB | Display | |
| Data in XML |  emd_10675_validation.xml.gz emd_10675_validation.xml.gz | 18.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10675 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10675 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10675 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10675 | HTTPS FTP |
-Related structure data
| Similar structure data | |
|---|---|
| EM raw data |  EMPIAR-10408 (Title: Single particle cryo-EM dataset of mouse heavy chain apoferritin collected on cryoARM300 EMPIAR-10408 (Title: Single particle cryo-EM dataset of mouse heavy chain apoferritin collected on cryoARM300Data size: 486.7 Data #1: raw uncorrected multiframe micrographs of mouse heavy chain apoferritin [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10675.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10675.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened masked map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.603 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10675_msk_1.map emd_10675_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: unfiltered half map 1
| File | emd_10675_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unfiltered half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: unfiltered half map 2
| File | emd_10675_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unfiltered half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : mouse heavy chain apoferritin
| Entire | Name: mouse heavy chain apoferritin |
|---|---|
| Components |
|
-Supramolecule #1: mouse heavy chain apoferritin
| Supramolecule | Name: mouse heavy chain apoferritin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Wilde type, octamer |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 506 KDa |
-Macromolecule #1: mouse heavy chain apoferritin
| Macromolecule | Name: mouse heavy chain apoferritin / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: ferroxidase |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MTTASPSQVR QNYHQDAEAA INRQINLELY ASYVYLSMSC YFDRDDVALK NFAKYFLHQS HEEREHAEK LMKLQNQRGG RIFLQDIKKP DRDDWESGLN AMECALHLEK SVNQSLLELH K LATDKNDP HLCDFIETYY LSEQVKSIKE LGDHVTNLRK MGAPEAGMAE YLFDKHTLGH GD ES |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 20 mM Hepes pH 7.5, 300 mM NaCl |
| Grid | Model: UltrAuFoil / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 98 % / Chamber temperature: 298 K / Instrument: GATAN CRYOPLUNGE 3 / Details: 4 seconds blotting. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Alignment procedure | Coma free - Residual tilt: 0.3 mrad |
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average exposure time: 8.0 sec. / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.55 mm |
| Sample stage | Specimen holder model: JEOL CRYOSPECPORTER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER / Target criteria: correlation coefficient |
 Movie
Movie Controller
Controller

































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)