[English] 日本語
 Yorodumi
Yorodumi- EMDB-10557: Cryo- EM structure of the Thermosynechococcus elongatus photosyst... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10557 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo- EM structure of the Thermosynechococcus elongatus photosystem I in the presence of cytochrome c6 | ||||||||||||
 Map data Map data | Structure of trimeric photosystem I from Thermosynechococcus elongatus. Asymmetric unit. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Photosystem I / P700 / Cyanobacteria / PHOTOSYNTHESIS | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationphotosystem I reaction center / photosystem I / photosynthetic electron transport in photosystem I / photosystem I / chlorophyll binding / plasma membrane-derived thylakoid membrane / photosynthesis / 4 iron, 4 sulfur cluster binding / electron transfer activity / oxidoreductase activity ...photosystem I reaction center / photosystem I / photosynthetic electron transport in photosystem I / photosystem I / chlorophyll binding / plasma membrane-derived thylakoid membrane / photosynthesis / 4 iron, 4 sulfur cluster binding / electron transfer activity / oxidoreductase activity / magnesium ion binding / metal ion binding / membrane Similarity search - Function | ||||||||||||
| Biological species |   Thermosynechococcus elongatus BP-1 (bacteria) / Thermosynechococcus elongatus BP-1 (bacteria) /   Thermosynechococcus elongatus (strain BP-1) (bacteria) Thermosynechococcus elongatus (strain BP-1) (bacteria) | ||||||||||||
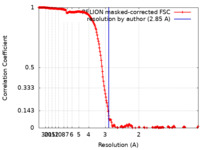
| Method | single particle reconstruction / cryo EM / Resolution: 2.85 Å | ||||||||||||
 Authors Authors | Koelsch A / Radon C | ||||||||||||
| Funding support |  Germany, 3 items Germany, 3 items
| ||||||||||||
 Citation Citation |  Journal: Curr.Opin.Struct.Biol. / Year: 2020 Journal: Curr.Opin.Struct.Biol. / Year: 2020Title: Current limits of structural biology: The transient interaction between cytochrome c6 and photosystem I Authors: Koelsch A / Radon C / Golub M / Baumert A / Burger J / Miehlke T / Lisdat F / Feoktystov A / Pieper J / Zouni A / Wendler P | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10557.map.gz emd_10557.map.gz | 37.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10557-v30.xml emd-10557-v30.xml emd-10557.xml emd-10557.xml | 27.1 KB 27.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10557_fsc.xml emd_10557_fsc.xml | 19.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_10557.png emd_10557.png | 51.7 KB | ||
| Filedesc metadata |  emd-10557.cif.gz emd-10557.cif.gz | 8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10557 http://ftp.pdbj.org/pub/emdb/structures/EMD-10557 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10557 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10557 | HTTPS FTP |
-Validation report
| Summary document |  emd_10557_validation.pdf.gz emd_10557_validation.pdf.gz | 385.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10557_full_validation.pdf.gz emd_10557_full_validation.pdf.gz | 384.8 KB | Display | |
| Data in XML |  emd_10557_validation.xml.gz emd_10557_validation.xml.gz | 17.1 KB | Display | |
| Data in CIF |  emd_10557_validation.cif.gz emd_10557_validation.cif.gz | 23.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10557 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10557 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10557 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10557 | HTTPS FTP |
-Related structure data
| Related structure data |  6traMC  6trcC  6trdC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10557.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10557.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of trimeric photosystem I from Thermosynechococcus elongatus. Asymmetric unit. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.628 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : photosystem I
+Supramolecule #1: photosystem I
+Macromolecule #1: Photosystem I P700 chlorophyll a apoprotein A1
+Macromolecule #2: Photosystem I P700 chlorophyll a apoprotein A2
+Macromolecule #3: Photosystem I iron-sulfur center
+Macromolecule #4: Photosystem I reaction center subunit II
+Macromolecule #5: Photosystem I reaction center subunit IV
+Macromolecule #6: Photosystem I reaction center subunit III
+Macromolecule #7: Photosystem I reaction center subunit VIII
+Macromolecule #8: Photosystem I reaction center subunit IX
+Macromolecule #9: Photosystem I reaction center subunit PsaK
+Macromolecule #10: Photosystem I reaction center subunit XI
+Macromolecule #11: Photosystem I reaction center subunit XII
+Macromolecule #12: Photosystem I 4.8K protein
+Macromolecule #13: CHLOROPHYLL A ISOMER
+Macromolecule #14: CHLOROPHYLL A
+Macromolecule #15: PHYLLOQUINONE
+Macromolecule #16: BETA-CAROTENE
+Macromolecule #17: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
+Macromolecule #18: IRON/SULFUR CLUSTER
+Macromolecule #19: CALCIUM ION
+Macromolecule #20: 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE
+Macromolecule #21: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE | ||||||||||||
| Details | Photosystem I was crosslinked with its electron donor cytochrome c6. Crosslinked particles are monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)