[English] 日本語
 Yorodumi
Yorodumi- EMDB-10553: Cryo-EM of native human uromodulin (UMOD)/Tamm-Horsfall protein (... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10553 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM of native human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament | |||||||||||||||
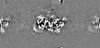
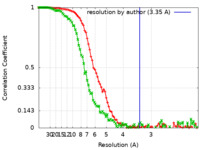
 Map data Map data | Density-modified cryo-em map of native uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament core (estimated resolution 3.35 A). | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | ZP MODULE / ZP DOMAIN / ZP-N DOMAIN / ZP-C DOMAIN / INTERDOMAIN LINKER / EGF DOMAIN / EXTRACELLULAR MATRIX / GLYCOPROTEIN / N-GLYCAN / STRUCTURAL PROTEIN / PROTEIN FILAMENT / PROTEIN POLYMERIZATION | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationcitric acid secretion / metanephric thick ascending limb development / micturition / metanephric distal convoluted tubule development / connective tissue replacement / protein transport into plasma membrane raft / organ or tissue specific immune response / Asparagine N-linked glycosylation / collecting duct development / urea transmembrane transport ...citric acid secretion / metanephric thick ascending limb development / micturition / metanephric distal convoluted tubule development / connective tissue replacement / protein transport into plasma membrane raft / organ or tissue specific immune response / Asparagine N-linked glycosylation / collecting duct development / urea transmembrane transport / metanephric ascending thin limb development / protein localization to vacuole / regulation of protein transport / juxtaglomerular apparatus development / antibacterial innate immune response / intracellular chloride ion homeostasis / renal urate salt excretion / glomerular filtration / urate transport / renal sodium ion absorption / neutrophil migration / response to water deprivation / intracellular phosphate ion homeostasis / potassium ion homeostasis / regulation of urine volume / intracellular sodium ion homeostasis / endoplasmic reticulum organization / IgG binding / heterophilic cell-cell adhesion / extrinsic component of membrane / ciliary membrane / leukocyte cell-cell adhesion / cellular response to unfolded protein / cellular defense response / multicellular organismal response to stress / renal water homeostasis / side of membrane / : / ERAD pathway / RNA splicing / tumor necrosis factor-mediated signaling pathway / apoptotic signaling pathway / autophagy / lipid metabolic process / Golgi lumen / regulation of blood pressure / intracellular calcium ion homeostasis / spindle pole / response to lipopolysaccharide / defense response to Gram-negative bacterium / basolateral plasma membrane / cilium / apical plasma membrane / response to xenobiotic stimulus / inflammatory response / negative regulation of cell population proliferation / calcium ion binding / cell surface / endoplasmic reticulum / extracellular space / extracellular exosome / membrane Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.35 Å | |||||||||||||||
 Authors Authors | Stsiapanava A / Xu C | |||||||||||||||
| Funding support |  Sweden, Sweden,  Singapore, 4 items Singapore, 4 items
| |||||||||||||||
 Citation Citation | Journal: Curr Top Dev Biol / Year: 2018 Title: Structure of Zona Pellucida Module Proteins. Authors: Marcel Bokhove / Luca Jovine /  Abstract: The egg coat, an extracellular matrix made up of glycoprotein filaments, plays a key role in animal fertilization by acting as a gatekeeper for sperm. Egg coat components polymerize using a common ...The egg coat, an extracellular matrix made up of glycoprotein filaments, plays a key role in animal fertilization by acting as a gatekeeper for sperm. Egg coat components polymerize using a common zona pellucida (ZP) "domain" module that consists of two related immunoglobulin-like domains, called ZP-N and ZP-C. The ZP module has also been recognized in a large number of other secreted proteins with different biological functions, whose mutations are linked to severe human diseases. During the last decade, tremendous progress has been made toward understanding the atomic architecture of the ZP module and the structural basis of its polymerization. Moreover, sperm-binding regions at the N-terminus of mollusk and mammalian egg coat subunits were found to consist of domain repeats that also adopt a ZP-N fold. This discovery revealed an unexpected link between invertebrate and vertebrate fertilization and led to the first structure of an egg coat-sperm protein recognition complex. In this review we summarize these exciting findings, discuss their functional implications, and outline future challenges that must be addressed in order to develop a comprehensive view of this family of biomedically important extracellular molecules. #2:  Journal: Biol. Cellulaire / Year: 1980 Journal: Biol. Cellulaire / Year: 1980Title: Etude chimique et ultrastructurale de la glycoproteine de Tamm et Horsfall ou uromucoide. Authors: Delain E / Thiery JP / Coulard D / Joliviene A / Hartman L #14:  Journal: bioRxiv / Year: 2020 Journal: bioRxiv / Year: 2020Title: Cryo-EM structure of native human uromodulin, a zona pellucida module polymer. Authors: Stsiapanava A / Xu C / Brunati M / Zamora-Caballero S / Schaeffer C / Han L / Carroni M / Yasumasu S / Rampoldi L / Wu B / Jovine L | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10553.map.gz emd_10553.map.gz | 7.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10553-v30.xml emd-10553-v30.xml emd-10553.xml emd-10553.xml | 41.2 KB 41.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10553_fsc.xml emd_10553_fsc.xml emd_10553_fsc_2.xml emd_10553_fsc_2.xml | 11.5 KB 12.4 KB | Display Display |  FSC data file FSC data file |
| Images |  emd_10553.png emd_10553.png | 72.6 KB | ||
| Filedesc metadata |  emd-10553.cif.gz emd-10553.cif.gz | 9.2 KB | ||
| Others |  emd_10553_additional_1.map.gz emd_10553_additional_1.map.gz emd_10553_additional_2.map.gz emd_10553_additional_2.map.gz emd_10553_half_map_1.map.gz emd_10553_half_map_1.map.gz emd_10553_half_map_2.map.gz emd_10553_half_map_2.map.gz | 12.4 MB 46 MB 46.2 MB 46.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10553 http://ftp.pdbj.org/pub/emdb/structures/EMD-10553 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10553 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10553 | HTTPS FTP |
-Related structure data
| Related structure data |  6tqkMC  6tqlC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10553.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10553.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density-modified cryo-em map of native uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament core (estimated resolution 3.35 A). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Density-modified cryo-em map of native uromodulin (UMOD)/Tamm-Horsfall protein...
| File | emd_10553_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
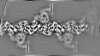
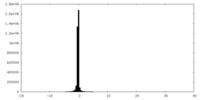
| Annotation | Density-modified cryo-em map of native uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament (estimated resolution 3.76 A). | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Unsharpened cryo-EM map of native uromodulin (UMOD)/Tamm-Horsfall protein...
| File | emd_10553_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
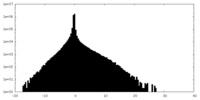
| Annotation | Unsharpened cryo-EM map of native uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unsharpened cryo-EM half map of native uromodulin (UMOD)/Tamm-Horsfall...
| File | emd_10553_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
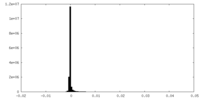
| Annotation | Unsharpened cryo-EM half map of native uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unsharpened cryo-EM half map of native uromodulin (UMOD)/Tamm-Horsfall...
| File | emd_10553_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened cryo-EM half map of native uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Uromodulin (UMOD)/Tamm-Horsfall protein (THP)
| Entire | Name: Uromodulin (UMOD)/Tamm-Horsfall protein (THP) |
|---|---|
| Components |
|
-Supramolecule #1: Uromodulin (UMOD)/Tamm-Horsfall protein (THP)
| Supramolecule | Name: Uromodulin (UMOD)/Tamm-Horsfall protein (THP) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Uromodulin
| Macromolecule | Name: Uromodulin / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 61.498816 KDa |
| Sequence | String: DTSEARWCSE CHSNATCTED EAVTTCTCQE GFTGDGLTCV DLDECAIPGA HNCSANSSCV NTPGSFSCVC PEGFRLSPGL GCTDVDECA EPGLSHCHAL ATCVNVVGSY LCVCPAGYRG DGWHCECSPG SCGPGLDCVP EGDALVCADP CQAHRTLDEY W RSTEYGEG ...String: DTSEARWCSE CHSNATCTED EAVTTCTCQE GFTGDGLTCV DLDECAIPGA HNCSANSSCV NTPGSFSCVC PEGFRLSPGL GCTDVDECA EPGLSHCHAL ATCVNVVGSY LCVCPAGYRG DGWHCECSPG SCGPGLDCVP EGDALVCADP CQAHRTLDEY W RSTEYGEG YACDTDLRGW YRFVGQGGAR MAETCVPVLR CNTAAPMWLN GTHPSSDEGI VSRKACAHWS GHCCLWDASV QV KACAGGY YVYNLTAPPE CHLAYCTDPS SVEGTCEECS IDEDCKSNNG RWHCQCKQDF NITDISLLEH RLECGANDMK VSL GKCQLK SLGFDKVFMY LSDSRCSGFN DRDNRDWVSV VTPARDGPCG TVLTRNETHA TYSNTLYLAD EIIIRDLNIK INFA CSYPL DMKVSLKTAL QPMVSALNIR VGGTGMFTVR MALFQTPSYT QPYQGSSVTL STEAFLYVGT MLDGGDLSRF ALLMT NCYA TPSSNATDPL KYFIIQDRCP HTRDSTIQVV ENGESSQGRF SVQMFRFAGN YDLVYLHCEV YLCDTMNEKC KPTCSG TRF UniProtKB: Uromodulin |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.85 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber temperature: 294 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 2300 / Average exposure time: 6.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 3.5 µm / Calibrated defocus min: 1.4000000000000001 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Overall B value: 115.88 | ||||||
| Output model |  PDB-6tqk: |
 Movie
Movie Controller
Controller

















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)