[English] 日本語
 Yorodumi
Yorodumi- EMDB-10359: Bat Influenza A polymerase product dissociation complex using 44-... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10359 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bat Influenza A polymerase product dissociation complex using 44-mer vRNA template with intact oligo(U) sequence | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Influenza / polymerase / viral transcription / RNA / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcap snatching / viral transcription / symbiont-mediated suppression of host mRNA transcription via inhibition of RNA polymerase II activity / 7-methylguanosine mRNA capping / host cell mitochondrion / virion component / host cell cytoplasm / symbiont-mediated suppression of host gene expression / viral translational frameshifting / RNA-directed RNA polymerase ...cap snatching / viral transcription / symbiont-mediated suppression of host mRNA transcription via inhibition of RNA polymerase II activity / 7-methylguanosine mRNA capping / host cell mitochondrion / virion component / host cell cytoplasm / symbiont-mediated suppression of host gene expression / viral translational frameshifting / RNA-directed RNA polymerase / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / GTP binding / host cell nucleus / RNA binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Influenza A virus (A/little yellow-shouldered bat/Guatemala/060/2010(H17N10)) / Influenza A virus (A/little yellow-shouldered bat/Guatemala/060/2010(H17N10)) /  Influenza B virus Influenza B virus | |||||||||
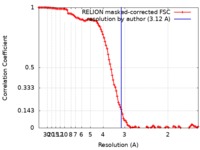
| Method | single particle reconstruction / cryo EM / Resolution: 3.12 Å | |||||||||
 Authors Authors | Wandzik JM / Kouba T | |||||||||
| Funding support |  France, 1 items France, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2020 Journal: Cell / Year: 2020Title: A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase. Authors: Joanna M Wandzik / Tomas Kouba / Manikandan Karuppasamy / Alexander Pflug / Petra Drncova / Jan Provaznik / Nayara Azevedo / Stephen Cusack /   Abstract: Influenza polymerase uses unique mechanisms to synthesize capped and polyadenylated mRNAs from the genomic viral RNA (vRNA) template, which is packaged inside ribonucleoprotein particles (vRNPs). ...Influenza polymerase uses unique mechanisms to synthesize capped and polyadenylated mRNAs from the genomic viral RNA (vRNA) template, which is packaged inside ribonucleoprotein particles (vRNPs). Here, we visualize by cryoelectron microscopy the conformational dynamics of the polymerase during the complete transcription cycle from pre-initiation to termination, focusing on the template trajectory. After exiting the active site cavity, the template 3' extremity rebinds into a specific site on the polymerase surface. Here, it remains sequestered during all subsequent transcription steps, forcing the template to loop out as it further translocates. At termination, the strained connection between the bound template 5' end and the active site results in polyadenylation by stuttering at uridine 17. Upon product dissociation, further conformational changes release the trapped template, allowing recycling back into the pre-initiation state. Influenza polymerase thus performs transcription while tightly binding to and protecting both template ends, allowing efficient production of multiple mRNAs from a single vRNP. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10359.map.gz emd_10359.map.gz | 108.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10359-v30.xml emd-10359-v30.xml emd-10359.xml emd-10359.xml | 26.1 KB 26.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10359_fsc.xml emd_10359_fsc.xml | 11.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_10359.png emd_10359.png | 133.6 KB | ||
| Filedesc metadata |  emd-10359.cif.gz emd-10359.cif.gz | 8 KB | ||
| Others |  emd_10359_additional.map.gz emd_10359_additional.map.gz emd_10359_half_map_1.map.gz emd_10359_half_map_1.map.gz emd_10359_half_map_2.map.gz emd_10359_half_map_2.map.gz | 55.5 MB 91.2 MB 91.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10359 http://ftp.pdbj.org/pub/emdb/structures/EMD-10359 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10359 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10359 | HTTPS FTP |
-Validation report
| Summary document |  emd_10359_validation.pdf.gz emd_10359_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10359_full_validation.pdf.gz emd_10359_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_10359_validation.xml.gz emd_10359_validation.xml.gz | 18.3 KB | Display | |
| Data in CIF |  emd_10359_validation.cif.gz emd_10359_validation.cif.gz | 24 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10359 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10359 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10359 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10359 | HTTPS FTP |
-Related structure data
| Related structure data |  6t0uMC  6szuC  6szvC  6t0nC  6t0rC  6t0sC  6t0vC  6t0wC  6t2cC  6tu5C  6tw1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10359.map.gz / Format: CCP4 / Size: 115.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10359.map.gz / Format: CCP4 / Size: 115.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
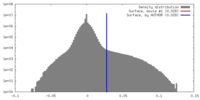
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: LocScale map
| File | emd_10359_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | LocScale map | ||||||||||||
| Projections & Slices |
| ||||||||||||
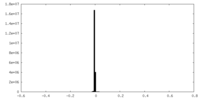
| Density Histograms |
-Half map: #1
| File | emd_10359_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
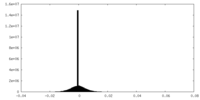
| Density Histograms |
-Half map: #2
| File | emd_10359_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
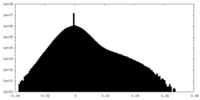
| Density Histograms |
- Sample components
Sample components
-Entire : Bat Influenza A polymerase product dissociation complex using 44-...
| Entire | Name: Bat Influenza A polymerase product dissociation complex using 44-mer vRNA template with intact oligo(U) sequence |
|---|---|
| Components |
|
-Supramolecule #1: Bat Influenza A polymerase product dissociation complex using 44-...
| Supramolecule | Name: Bat Influenza A polymerase product dissociation complex using 44-mer vRNA template with intact oligo(U) sequence type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: Polymerase
| Supramolecule | Name: Polymerase / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/little yellow-shouldered bat/Guatemala/060/2010(H17N10)) Influenza A virus (A/little yellow-shouldered bat/Guatemala/060/2010(H17N10)) |
-Supramolecule #3: Nucleic acid
| Supramolecule | Name: Nucleic acid / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #4 |
|---|---|
| Source (natural) | Organism:  Influenza B virus Influenza B virus |
-Macromolecule #1: Polymerase acidic protein
| Macromolecule | Name: Polymerase acidic protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/little yellow-shouldered bat/Guatemala/060/2010(H17N10)) Influenza A virus (A/little yellow-shouldered bat/Guatemala/060/2010(H17N10)) |
| Molecular weight | Theoretical: 85.432828 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GSHHHHHHHH GSGSMENFVR TNFNPMILER AEKTMKEYGE NPQNEGNKFA AISTHMEVCF MYSDFHFIDL EGNTIVKEND DDNAMLKHR FEIIEGQERN IAWTIVNSIC NMTENSKPRF LPDLYDYKTN KFIEIGVTRR KVEDYYYEAA SKLKGENVYI H IFSFDGEE ...String: GSHHHHHHHH GSGSMENFVR TNFNPMILER AEKTMKEYGE NPQNEGNKFA AISTHMEVCF MYSDFHFIDL EGNTIVKEND DDNAMLKHR FEIIEGQERN IAWTIVNSIC NMTENSKPRF LPDLYDYKTN KFIEIGVTRR KVEDYYYEAA SKLKGENVYI H IFSFDGEE MATDDEYILD EESRARIKTR LFVLRQELAT AGLWDSFRQS EKGEETLEEE FSYPPTFQRL ANQSLPPSFK DY HQFKAYV SSFKANGNIE AKLGAMSEKV NAQIESFDPR TIRELELPEG KFCTQRSKFL LMDAMKLSVL NPAHEGEGIP MKD AKACLD TFWGWKKATI IKKHEKGVNT NYLMIWEQLL ESIKEMEGKF LNLKKTNHLK WGLGEGQAPE KMDFEDCKEV PDLF QYKSE PPEKRKLASW IQSEFNKASE LTNSNWIEFD ELGNDVAPIE HIASRRRNFF TAEVSQCRAS EYIMKAVYIN TALLN SSCT AMEEYQVIPI ITKCRDTSGQ RRTNLYGFII KGRSHLRNDT DVVNFISLEF SLTDPRNEIH KWEKYCVLEI GDMEIR TSI STIMKPVYLY VRTNGTSKIK MKWGMEMRRC LLQSLQQVES MIEAESAVKE KDMTEPFFRN RENDWPIGES PQGIEKG TI GKVCRVLLAK SVFNSIYASA QLEGFSAESR KLLLLIQAFR DNLDPGTFDL KGLYEAIEEC IINDPWVLLN ASWFNSFL K AVQLSMGSGS GENLYFQ UniProtKB: Polymerase acidic protein |
-Macromolecule #2: RNA-directed RNA polymerase catalytic subunit
| Macromolecule | Name: RNA-directed RNA polymerase catalytic subunit / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/little yellow-shouldered bat/Guatemala/060/2010(H17N10)) Influenza A virus (A/little yellow-shouldered bat/Guatemala/060/2010(H17N10)) |
| Molecular weight | Theoretical: 87.936312 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GSGSGSGSGM DVNPMLIFLK VPVQNAISTT FPYTGDPPYS HGTGTGYTMD TVIRTHDYSS RGIWKTNSET GAQQLNPIDG PLPEDNEPS GYAQTDCVLE LIEGLDRSHP GLFETACQET IDAIQQTRVD KLTQGRQTYD WTLNRNQPAA TALANTIEVF R KNGYKLNE ...String: GSGSGSGSGM DVNPMLIFLK VPVQNAISTT FPYTGDPPYS HGTGTGYTMD TVIRTHDYSS RGIWKTNSET GAQQLNPIDG PLPEDNEPS GYAQTDCVLE LIEGLDRSHP GLFETACQET IDAIQQTRVD KLTQGRQTYD WTLNRNQPAA TALANTIEVF R KNGYKLNE SGRLIDFLKD VLLSFENDSM EVTTHFQKKK RIRDNHSKKM ITQRTIGKKR VKLTKKNYLI RALTLNTMTK DA ERGKLKR RAIATPGMQI RGFVYFVELL ARNICERLEQ SGLPVGGNEK KAKLANVIKK MMAKSTDEEL SYTITGDNTK WNE NQNPRI FLAMVLRITA GQPEWFRDLL AVAPIMFSNK VARLGRGYMF ESKSMHLRTQ ISAENLSDIN LRYFNEDTKK KIEK IRHLM VEGTASLSPG MMMGMFNMLS TVLGVSVLNL GQREILKRTY WWDGLQSSDD FALIINGHFK EDIQQGVNHF YRTCK LVGI NMSQKKSYIN KTGTFEFTSF FYRYGFVANF SMELPSFGVA GNNESADMSI GTTVIKTNMI NNDLGPATAQ MAIQLF IKD YRYTYRCHRG DTNLETRRTK SIKRLWTETI SKAGLLVADG GPNPYNLRNL HIPEVCLKWS LMDPDYRGRL CNPNNPF VH HMEVESTNLA VVMPAHGPAK SLEYDAVATT HSWTPKRNRS ILNTNQRGIL EDERIYQKCC QVFEKFFPSS TYRRPIGM A SMLDAMLSRA RIDARIDLES GRISSQDFSE ITNTCKAIEA LKRQGSGSGE NLYFQ UniProtKB: RNA-directed RNA polymerase catalytic subunit |
-Macromolecule #3: Polymerase basic protein 2
| Macromolecule | Name: Polymerase basic protein 2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Influenza A virus (A/little yellow-shouldered bat/Guatemala/060/2010(H17N10)) Influenza A virus (A/little yellow-shouldered bat/Guatemala/060/2010(H17N10)) |
| Molecular weight | Theoretical: 91.027141 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GSGSGSGSGM ERIKELMEMV KNSRMREILT TTSVDHMAVI KKYTSGRQEK NPALRMKWMM AMKYPISASS RIREMIPEKD EDGNTLWTN TKDAGSNRVL VSPNAVTWWN RAGPVSDVVH YPRVYKMYFD RLERLTHGTF GPVKFYNQVK VRKRVDINPG H KDLTSREA ...String: GSGSGSGSGM ERIKELMEMV KNSRMREILT TTSVDHMAVI KKYTSGRQEK NPALRMKWMM AMKYPISASS RIREMIPEKD EDGNTLWTN TKDAGSNRVL VSPNAVTWWN RAGPVSDVVH YPRVYKMYFD RLERLTHGTF GPVKFYNQVK VRKRVDINPG H KDLTSREA QEVIMEVVFP NEVGARTLSS DAQLTITKEK KEELKNCKIS PIMVAYMLER ELVRRTRFLP IAGATSSTYV EV LHLTQGT CWEQQYTPGG EAENDDLDQT LIIASRNIVR RSIVAIDPLA SLLSMCHTTS ISSEPLVEIL RSNPTDEQAV NIC KAALGI RINNSFSFGG YNFKRVKGSS QRTEKAVLTG NLQTLTMTIF EGYEEFNVSG KRASAVLKKG AQRLIQAIIG GRTL EDILN LMITLMVFSQ EEKMLKAVRG DLNFVNRANQ RLNPMYQLLR HFQKDSSTLL KNWGTEEIDP IMGIAGIMPD GTINK TQTL MGVRLSQGGV DEYSFNERIR VNIDKYLRVR NEKGELLISP EEVSEAQGQE KLPINYNSSL MWEVNGPESI LTNTYH WII KNWELLKTQW MTDPTVLYNR IEFEPFQTLI PKGNRAIYSG FTRTLFQQMR DVEGTFDSIQ IIKLLPFSAH PPSLGRT QF SSFTLNIRGA PLRLLIRGNS QVFNYNQMEN VIIVLGKSVG SPERSILTES SSIESAVLRG FLILGKANSK YGPVLTIG E LDKLGRGEKA NVLIGQGDTV LVMKRKRDSS ILTDSQTALK RIRLEESKGW SHPQFEKGGG SGGGSGGSAW SHPQFEKGR SGGENLYFQ UniProtKB: Polymerase basic protein 2 |
-Macromolecule #4: vRNA
| Macromolecule | Name: vRNA / type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Influenza B virus Influenza B virus |
| Molecular weight | Theoretical: 13.911169 KDa |
| Sequence | String: AGUAGUAACA AGAGCAUUUU UUCCGUCUCG CCUCUGCUUC UGCU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 43.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)