+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10045 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
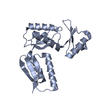
| Title | Inner membrane ring of the Shigella type 3 secretion system | |||||||||
 Map data Map data | Masked C24 map of inner membrane ring of Shigella type 3 secretion system, post-processed, sharpened (b=-128) and filtered at 3.55 A. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | type 3 secretion system / shigella / ring-forming membrane protein / PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Shigella flexneri (bacteria) Shigella flexneri (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.55 Å | |||||||||
 Authors Authors | Lunelli M / Kamprad A | |||||||||
| Funding support | 2 items
| |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2020 Journal: PLoS Pathog / Year: 2020Title: Cryo-EM structure of the Shigella type III needle complex. Authors: Michele Lunelli / Antje Kamprad / Jörg Bürger / Thorsten Mielke / Christian M T Spahn / Michael Kolbe /  Abstract: The Type III Secretion Systems (T3SS) needle complex is a conserved syringe-shaped protein translocation nanomachine with a mass of about 3.5 MDa essential for the survival and virulence of many Gram- ...The Type III Secretion Systems (T3SS) needle complex is a conserved syringe-shaped protein translocation nanomachine with a mass of about 3.5 MDa essential for the survival and virulence of many Gram-negative bacterial pathogens. This system is composed of a membrane-embedded basal body and an extracellular needle that deliver effector proteins into host cells. High-resolution structures of the T3SS from different organisms and infection stages are needed to understand the underlying molecular mechanisms of effector translocation. Here, we present the cryo-electron microscopy structure of the isolated Shigella T3SS needle complex. The inner membrane (IM) region of the basal body adopts 24-fold rotational symmetry and forms a channel system that connects the bacterial periplasm with the export apparatus cage. The secretin oligomer adopts a heterogeneous architecture with 16- and 15-fold cyclic symmetry in the periplasmic N-terminal connector and C-terminal outer membrane ring, respectively. Two out of three IM subunits bind the secretin connector via a β-sheet augmentation. The cryo-EM map also reveals the helical architecture of the export apparatus core, the inner rod, the needle and their intervening interfaces. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10045.map.gz emd_10045.map.gz | 31.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10045-v30.xml emd-10045-v30.xml emd-10045.xml emd-10045.xml | 22.6 KB 22.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10045_fsc.xml emd_10045_fsc.xml | 16.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_10045.png emd_10045.png | 87.9 KB | ||
| Masks |  emd_10045_msk_1.map emd_10045_msk_1.map | 421.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10045.cif.gz emd-10045.cif.gz | 6.4 KB | ||
| Others |  emd_10045_additional.map.gz emd_10045_additional.map.gz emd_10045_half_map_1.map.gz emd_10045_half_map_1.map.gz emd_10045_half_map_2.map.gz emd_10045_half_map_2.map.gz | 32.5 MB 331.3 MB 330 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10045 http://ftp.pdbj.org/pub/emdb/structures/EMD-10045 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10045 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10045 | HTTPS FTP |
-Related structure data
| Related structure data |  6rwxMC  6rwkC  6rwyC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10045.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10045.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Masked C24 map of inner membrane ring of Shigella type 3 secretion system, post-processed, sharpened (b=-128) and filtered at 3.55 A. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.38369 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10045_msk_1.map emd_10045_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Masked C24 map of inner membrane ring of...
| File | emd_10045_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Masked C24 map of inner membrane ring of Shigella type 3 secretion system, post-processed, sharpened (b=-128) and filtered at 3.2 A. Used for model building and refinement. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unfiltered half map
| File | emd_10045_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unfiltered half map
| File | emd_10045_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The periplasmic inner membrane ring of the Shigella type 3 secret...
| Entire | Name: The periplasmic inner membrane ring of the Shigella type 3 secretion system |
|---|---|
| Components |
|
-Supramolecule #1: The periplasmic inner membrane ring of the Shigella type 3 secret...
| Supramolecule | Name: The periplasmic inner membrane ring of the Shigella type 3 secretion system type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Focused reconstruction from isolated needle complexes of the Shigella type 3 secretion system |
|---|---|
| Source (natural) | Organism:  Shigella flexneri (bacteria) / Strain: M90T / Location in cell: Membrane Shigella flexneri (bacteria) / Strain: M90T / Location in cell: Membrane |
-Macromolecule #1: Protein MxiG
| Macromolecule | Name: Protein MxiG / type: protein_or_peptide / ID: 1 / Number of copies: 24 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Shigella flexneri (bacteria) Shigella flexneri (bacteria) |
| Molecular weight | Theoretical: 43.053844 KDa |
| Sequence | String: MSEAKNSNLA PFRLLVKLTN GVGDEFPLYY GNNLIVLGRT IETLEFGNDN FPENIIPVTD SKSDGIIYLT ISKDNICQFS DEKGEQIDI NSQFNSFEYD GISFHLKNMR EDKSRGHILN GMYKNHSVFF FFAVIVVLII IFSLSLKKDE VKEIAEIIDD K RYGIVNTG ...String: MSEAKNSNLA PFRLLVKLTN GVGDEFPLYY GNNLIVLGRT IETLEFGNDN FPENIIPVTD SKSDGIIYLT ISKDNICQFS DEKGEQIDI NSQFNSFEYD GISFHLKNMR EDKSRGHILN GMYKNHSVFF FFAVIVVLII IFSLSLKKDE VKEIAEIIDD K RYGIVNTG QCNYILAETQ NDAVWASVAL NKTGFTKCRY ILVSNKEINR IQQYINQRFP FINLYVLNLV SDKAELLVFL SK ERNSSKD TELDKLKNAL IVEFPYIKNI KFNYLSDHNA RGDAKGIFTK VNVQYKEICE NNKVTYSVRE ELTDEKLELI NRL ISEHKN IYGDQYIEFS VLLIDDDFKG KSYLNSKDSY VMLNDKHWFF LDKNK UniProtKB: Protein MxiG |
-Macromolecule #2: Lipoprotein MxiJ
| Macromolecule | Name: Lipoprotein MxiJ / type: protein_or_peptide / ID: 2 / Number of copies: 24 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Shigella flexneri (bacteria) Shigella flexneri (bacteria) |
| Molecular weight | Theoretical: 27.542055 KDa |
| Sequence | String: MIRYKGFILF LLLMLIGCEQ REELISNLSQ RQANEIISVL ERHNITARKV DGGKQGISVQ VEKGTFASAV DLMRMYDLPN PERVDISQM FPTDSLVSSP RAEKARLYSA IEQRLEQSLV SIGGVISAKI HVSYDLEEKN ISSKPMHISV IAIYDSPKES E LLVSNIKR ...String: MIRYKGFILF LLLMLIGCEQ REELISNLSQ RQANEIISVL ERHNITARKV DGGKQGISVQ VEKGTFASAV DLMRMYDLPN PERVDISQM FPTDSLVSSP RAEKARLYSA IEQRLEQSLV SIGGVISAKI HVSYDLEEKN ISSKPMHISV IAIYDSPKES E LLVSNIKR FLKNTFSDVK YENISVILTP KEEYVYTNVQ PVKEVKSEFL TNEVIYLFLG MAVLVVILLV WAFKTGWFKR NK I UniProtKB: Lipoprotein MxiJ |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 5 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV Details: Sample applied on grid 5 ul, incubation time 5 min on ice, then moved into Vitrobot and 5 ul sample applied again. Blot time: 2 sec Blot force: -2 Drain time: 0 sec. |
| Details | Isolated needle complex in detergent solution |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 5238 / Average exposure time: 1.5 sec. / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 0.004 µm / Nominal defocus min: 0.0015 µm / Nominal magnification: 101179 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Details | Model was built and refined in the map low-pass filtered at 3.2 A | ||||||
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 140 / Target criteria: Correlation coefficient and geometry | ||||||
| Output model |  PDB-6rwx: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)