[English] 日本語
 Yorodumi
Yorodumi- EMDB-10561: Structure of the RBM3/collar region of the Salmonella flagella MS... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10561 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the RBM3/collar region of the Salmonella flagella MS-ring protein FliF with 35-fold symmetry applied | |||||||||||||||||||||
 Map data Map data | Post process map | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
| Biological species |  Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) | |||||||||||||||||||||
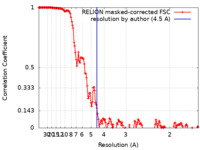
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||||||||||||||
 Authors Authors | Johnson S / Fong YH / Deme JC / Furlong EJ / Kuhlen L / Lea SM | |||||||||||||||||||||
| Funding support |  United Kingdom, 6 items United Kingdom, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2020 Journal: Nat Microbiol / Year: 2020Title: Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation. Authors: Steven Johnson / Yu Hang Fong / Justin C Deme / Emily J Furlong / Lucas Kuhlen / Susan M Lea /  Abstract: The bacterial flagellum is a complex self-assembling nanomachine that confers motility to the cell. Despite great variation across species, all flagella are ultimately constructed from a helical ...The bacterial flagellum is a complex self-assembling nanomachine that confers motility to the cell. Despite great variation across species, all flagella are ultimately constructed from a helical propeller that is attached to a motor embedded in the inner membrane. The motor consists of a series of stator units surrounding a central rotor made up of two ring complexes, the MS-ring and the C-ring. Despite many studies, high-resolution structural information is still lacking for the MS-ring of the rotor, and proposed mismatches in stoichiometry between the two rings have long provided a source of confusion for the field. Here, we present structures of the Salmonella MS-ring, revealing a high level of variation in inter- and intrachain symmetry that provides a structural explanation for the ability of the MS-ring to function as a complex and elegant interface between the two main functions of the flagellum-protein secretion and rotation. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10561.map.gz emd_10561.map.gz | 36.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10561-v30.xml emd-10561-v30.xml emd-10561.xml emd-10561.xml | 18.9 KB 18.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10561_fsc.xml emd_10561_fsc.xml | 15.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_10561.png emd_10561.png | 46.7 KB | ||
| Masks |  emd_10561_msk_1.map emd_10561_msk_1.map | 307.5 MB |  Mask map Mask map | |
| Others |  emd_10561_additional.map.gz emd_10561_additional.map.gz emd_10561_additional_1.map.gz emd_10561_additional_1.map.gz emd_10561_half_map_1.map.gz emd_10561_half_map_1.map.gz emd_10561_half_map_2.map.gz emd_10561_half_map_2.map.gz | 237.9 MB 237.9 MB 243.5 MB 243.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10561 http://ftp.pdbj.org/pub/emdb/structures/EMD-10561 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10561 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10561 | HTTPS FTP |
-Related structure data
| Related structure data |  6scnC  6sd1C  6sd2C  6sd3C  6sd4C 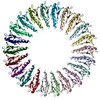 6sd5C  6treC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10561.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10561.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post process map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.822 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10561_msk_1.map emd_10561_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Refinement map
| File | emd_10561_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refinement map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Refinement map
| File | emd_10561_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refinement map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_10561_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_10561_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homomeric 35mer of Salmonella enterica serovar Typhimurium FliF
| Entire | Name: Homomeric 35mer of Salmonella enterica serovar Typhimurium FliF |
|---|---|
| Components |
|
-Supramolecule #1: Homomeric 35mer of Salmonella enterica serovar Typhimurium FliF
| Supramolecule | Name: Homomeric 35mer of Salmonella enterica serovar Typhimurium FliF type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 2.14 MDa |
-Macromolecule #1: Flagellar MS-ring FliF
| Macromolecule | Name: Flagellar MS-ring FliF / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MSATASTATQ PKPLEWLNRL RANPRIPLIV AGSAAVAIVV AMVLWAKTPD YRTLFSNLSD QDGGAIVAQ LTQMNIPYRF ANGSGAIEVP ADKVHELRLR LAQQGLPKGG AVGFELLDQE K FGISQFSE QVNYQRALEG ELARTIETLG PVKSARVHLA MPKPSLFVRE ...String: MSATASTATQ PKPLEWLNRL RANPRIPLIV AGSAAVAIVV AMVLWAKTPD YRTLFSNLSD QDGGAIVAQ LTQMNIPYRF ANGSGAIEVP ADKVHELRLR LAQQGLPKGG AVGFELLDQE K FGISQFSE QVNYQRALEG ELARTIETLG PVKSARVHLA MPKPSLFVRE QKSPSASVTV TL EPGRALD EGQISAVVHL VSSAVAGLPP GNVTLVDQSG HLLTQSNTSG RDLNDAQLKF AND VESRIQ RRIEAILSPI VGNGNVHAQV TAQLDFANKE QTEEHYSPNG DASKATLRSR QLNI SEQVG AGYPGGVPGA LSNQPAPPNE APIATPPTNQ QNAQNTPQTS TSTNSNSAGP RSTQR NETS NYEVDRTIRH TKMNVGDIER LSVAVVVNYK TLADGKPLPL TADQMKQIED LTREAM GFS DKRGDTLNVV NSPFSAVDNT GGELPFWQQQ SFIDQLLAAG RWLLVLVVAW ILWRKAV RP QLTRRVEEAK AAQEQAQVRQ ETEEAVEVRL SKDEQLQQRR ANQRLGAEVM SQRIREMS D NDPRVVALVI RQWMSNDHE |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)