[English] 日本語
 Yorodumi
Yorodumi- EMDB-0774: Rat GluD1 receptor(compact conformation) in complex with 7-CKA an... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0774 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Rat GluD1 receptor(compact conformation) in complex with 7-CKA and Calcium ions | |||||||||
 Map data Map data | final refined map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationGABA receptor activity / trans-synaptic protein complex / negative regulation of synaptic plasticity / G protein-coupled receptor activity involved in regulation of postsynaptic membrane potential / synaptic signaling via neuropeptide / regulation of postsynapse organization / AMPA glutamate receptor activity / AMPA glutamate receptor complex / social behavior / regulation of postsynaptic membrane neurotransmitter receptor levels ...GABA receptor activity / trans-synaptic protein complex / negative regulation of synaptic plasticity / G protein-coupled receptor activity involved in regulation of postsynaptic membrane potential / synaptic signaling via neuropeptide / regulation of postsynapse organization / AMPA glutamate receptor activity / AMPA glutamate receptor complex / social behavior / regulation of postsynaptic membrane neurotransmitter receptor levels / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / synaptic transmission, glutamatergic / postsynaptic density membrane / modulation of chemical synaptic transmission / GABA-ergic synapse / phospholipase C-activating G protein-coupled receptor signaling pathway / dendritic spine / postsynaptic membrane / glutamatergic synapse / metal ion binding / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.1 Å | |||||||||
 Authors Authors | Burada AP / Kumar J | |||||||||
| Funding support |  India, 1 items India, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2020 Journal: Nat Struct Mol Biol / Year: 2020Title: Cryo-EM structures of the ionotropic glutamate receptor GluD1 reveal a non-swapped architecture. Authors: Ananth Prasad Burada / Rajesh Vinnakota / Janesh Kumar /  Abstract: Ionotropic orphan delta (GluD) receptors are not gated by glutamate or any other endogenous ligand but are grouped with ionotropic glutamate receptors (iGluRs) based on sequence similarity. GluD1 ...Ionotropic orphan delta (GluD) receptors are not gated by glutamate or any other endogenous ligand but are grouped with ionotropic glutamate receptors (iGluRs) based on sequence similarity. GluD1 receptors play critical roles in synaptogenesis and synapse maintenance and have been implicated in neuronal disorders, including schizophrenia, cognitive deficits, and cerebral ataxia. Here we report cryo-EM structures of the rat GluD1 receptor complexed with calcium and the ligand 7-chlorokynurenic acid (7-CKA), elucidating molecular architecture and principles of receptor assembly. The structures reveal a non-swapped architecture at the interface of the extracellular amino-terminal domain (ATD) and the ligand-binding domain (LBD). This finding is in contrast with structures of other families of iGluRs, where the dimer partners between the ATD and LBD layers are swapped. Our results demonstrate that principles of architecture and symmetry are not conserved between delta receptors and other iGluRs and provide a molecular blueprint for understanding the functions of the 'orphan' class of iGluRs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0774.map.gz emd_0774.map.gz | 226.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0774-v30.xml emd-0774-v30.xml emd-0774.xml emd-0774.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0774.png emd_0774.png | 41.1 KB | ||
| Filedesc metadata |  emd-0774.cif.gz emd-0774.cif.gz | 6.1 KB | ||
| Others |  emd_0774_half_map_1.map.gz emd_0774_half_map_1.map.gz emd_0774_half_map_2.map.gz emd_0774_half_map_2.map.gz | 226.8 MB 226.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0774 http://ftp.pdbj.org/pub/emdb/structures/EMD-0774 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0774 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0774 | HTTPS FTP |
-Validation report
| Summary document |  emd_0774_validation.pdf.gz emd_0774_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_0774_full_validation.pdf.gz emd_0774_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_0774_validation.xml.gz emd_0774_validation.xml.gz | 15.8 KB | Display | |
| Data in CIF |  emd_0774_validation.cif.gz emd_0774_validation.cif.gz | 18.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0774 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0774 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0774 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0774 | HTTPS FTP |
-Related structure data
| Related structure data |  6kssMC  0773C  6kspC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10428 (Title: Cryo-EM Structure of GluD1-Orphan Delta Receptor Reveals a Novel Architecture in the Ionotropic Glutamate Receptor Family EMPIAR-10428 (Title: Cryo-EM Structure of GluD1-Orphan Delta Receptor Reveals a Novel Architecture in the Ionotropic Glutamate Receptor FamilyData size: 6.5 TB Data #1: ionotropioc Glutamate receptor Delta1 [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0774.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0774.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | final refined map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.067 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
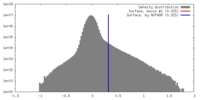
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: half map A
| File | emd_0774_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
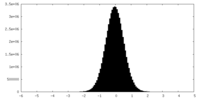
| Density Histograms |
-Half map: half map B
| File | emd_0774_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
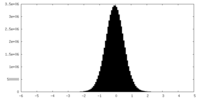
| Density Histograms |
- Sample components
Sample components
-Entire : GluD1
| Entire | Name: GluD1 |
|---|---|
| Components |
|
-Supramolecule #1: GluD1
| Supramolecule | Name: GluD1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Recombinantly expressed protein |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 388 KDa |
-Macromolecule #1: Glutamate receptor ionotropic, delta-1
| Macromolecule | Name: Glutamate receptor ionotropic, delta-1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 95.718039 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DSIIHIGAIF EENAAKDDRV FQLAVSDLSL NDDILQSEKI TYSIKVIEAN NPFQAVQEAC DLMTQGILAL VTSTGCASAN ALQSLTDAM HIPHLFVQRN PGGSPRTACH LNPSPDGEAY TLASRPPVRL NDVMLRLVTE LRWQKFVMFY DSEYDIRGLQ S FLDQASRL ...String: DSIIHIGAIF EENAAKDDRV FQLAVSDLSL NDDILQSEKI TYSIKVIEAN NPFQAVQEAC DLMTQGILAL VTSTGCASAN ALQSLTDAM HIPHLFVQRN PGGSPRTACH LNPSPDGEAY TLASRPPVRL NDVMLRLVTE LRWQKFVMFY DSEYDIRGLQ S FLDQASRL GLDVSLQKVD KNISHVFTSL FTTMKTEELN RYRDTLRRAI LLLSPQGAHS FINEAVETNL ASKDSHWVFV NE EISDPEI LDLVHSALGR MTVVRQIFPS AKDNQKCMRN NHRISSLLCD PQEGYLQMLQ ISNLYLYDSV LMLANAFHRK LED RKWHSM ASLNCIRKST KPWNGGRSML DTIKKGHITG LTGVMEFRED SSNPYVQFEI LGTTYSETFG KDMRKLATWD SEKG LNGSL QERPMGSRLQ GLTLKVVTVL EEPFVMVAEN ILGQPKRYKG FSIDVLDALA KALGFKYEIY QAPDGRYGHQ LHNTS WNGM IGELISKRAD LAISAITITP ERESVVDFSK RYMDYSVGIL IKKPEEKISI FSLFAPFDFA VWACIAAAIP VVGVLI FVL NRIQAVRSQS ATQPRPSASA TLHSAIWIVY GAFVQQGGES SVNSVAMRIV MGSWWLFTLI VCSSYTANLA AFLTVSR MD SPVRTFQDLS KQLEMSYGTV RDSAVYEYFR AKGTNPLEQD STFAELWRTI SKNGGADNCV SNPSEGIRKA KKGNYAFL W DVAVVEYAAL TDDDCSVTVI GNSISSKGYG IALQHGSPYR DLFSQRILEL QDTGDLDVLK QKWWPHTGRC DLTSHSSAQ TDGKSLKLHS FAGVFCILAI GLLLACLVAA LELWWNSNRC HQETPKEDKE VGLVPR UniProtKB: Glutamate receptor ionotropic, delta-1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.9 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 20mM Tris, 150mM NaCl, 0.75mM DDM, 0.025mM CHS |
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 0-40 / Number real images: 4120 / Average exposure time: 6.0 sec. / Average electron dose: 40.38 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6kss: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)