+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0716 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Hyperthermophilic respiratory Complex III | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Respiratory chain / Complex III / Hyperthermophilic mechanism / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationquinol-cytochrome-c reductase activity / respiratory electron transport chain / 2 iron, 2 sulfur cluster binding / oxidoreductase activity / electron transfer activity / heme binding / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   Aquifex aeolicus (bacteria) / Aquifex aeolicus (bacteria) /   Aquifex aeolicus (strain VF5) (bacteria) Aquifex aeolicus (strain VF5) (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Fei S / Hartmut M | |||||||||
 Citation Citation |  Journal: Angew Chem Int Ed Engl / Year: 2020 Journal: Angew Chem Int Ed Engl / Year: 2020Title: A 3.3 Å-Resolution Structure of Hyperthermophilic Respiratory Complex III Reveals the Mechanism of Its Thermal Stability. Authors: Guoliang Zhu / Hui Zeng / Shuangbo Zhang / Jana Juli / Xiaoyun Pang / Jan Hoffmann / Yan Zhang / Nina Morgner / Yun Zhu / Guohong Peng / Hartmut Michel / Fei Sun /   Abstract: Respiratory chain complexes convert energy by coupling electron flow to transmembrane proton translocation. Owing to a lack of atomic structures of cytochrome bc complex (Complex III) from ...Respiratory chain complexes convert energy by coupling electron flow to transmembrane proton translocation. Owing to a lack of atomic structures of cytochrome bc complex (Complex III) from thermophilic bacteria, little is known about the adaptations of this macromolecular machine to hyperthermophilic environments. In this study, we purified the cytochrome bc complex of Aquifex aeolicus, one of the most extreme thermophilic bacteria known, and determined its structure with and without an inhibitor at 3.3 Å resolution. Several residues unique for thermophilic bacteria were detected that provide additional stabilization for the structure. An extra transmembrane helix at the N-terminus of cyt. c was found to greatly enhance the interaction between cyt. b and cyt. c , and to bind a phospholipid molecule to stabilize the complex in the membrane. These results provide the structural basis for the hyperstability of the cytochrome bc complex in an extreme thermal environment. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0716.map.gz emd_0716.map.gz | 5.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0716-v30.xml emd-0716-v30.xml emd-0716.xml emd-0716.xml | 24.7 KB 24.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0716.png emd_0716.png | 90.7 KB | ||
| Filedesc metadata |  emd-0716.cif.gz emd-0716.cif.gz | 6.7 KB | ||
| Others |  emd_0716_half_map_1.map.gz emd_0716_half_map_1.map.gz emd_0716_half_map_2.map.gz emd_0716_half_map_2.map.gz | 48.5 MB 48.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0716 http://ftp.pdbj.org/pub/emdb/structures/EMD-0716 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0716 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0716 | HTTPS FTP |
-Related structure data
| Related structure data |  6klsMC  0719C  6klvC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0716.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0716.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #2
| File | emd_0716_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_0716_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : The cytochrome bc1 complex (respiratory complex III)
| Entire | Name: The cytochrome bc1 complex (respiratory complex III) |
|---|---|
| Components |
|
-Supramolecule #1: The cytochrome bc1 complex (respiratory complex III)
| Supramolecule | Name: The cytochrome bc1 complex (respiratory complex III) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Aquifex aeolicus (bacteria) Aquifex aeolicus (bacteria) |
-Macromolecule #1: Rieske-I iron sulfur protein
| Macromolecule | Name: Rieske-I iron sulfur protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Aquifex aeolicus (strain VF5) (bacteria) / Strain: VF5 Aquifex aeolicus (strain VF5) (bacteria) / Strain: VF5 |
| Molecular weight | Theoretical: 19.429857 KDa |
| Sequence | String: MEASRRDFIS IGIGALGAVG GLGALYALVR VMLEPSEIAA LGAKTEIDVS KIQPMQVRVT SWKGKTLFAI RLPKDFKPEG YTLKKGALN SKGTTNYEIL KGHDVFALVG VCTHLGCIPL WKPQGEGGIN KPVFHCPCHG GLYTPYGDVI GGPPPRPLFI P PQKLEGNK LIVGVEGFVK ELI UniProtKB: Rieske-I iron sulfur protein |
-Macromolecule #2: cytochrome b subunit
| Macromolecule | Name: cytochrome b subunit / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Aquifex aeolicus (bacteria) Aquifex aeolicus (bacteria) |
| Molecular weight | Theoretical: 47.045629 KDa |
| Sequence | String: MGLIEKIVDW IDERAHVREI YRTQMVEYKV AKNLTFPYVF GILALVTFAI QIISGMVLIL YYKPSIADAF DSATYSIMGE IPFGWLFRH IHATGANFFM AIVYLHMFTG IYYNAYKRPR ELVWIVGWLI YFVLILTALS GYLLPWGQLS YWGFIVTTEI P GSLADAPI ...String: MGLIEKIVDW IDERAHVREI YRTQMVEYKV AKNLTFPYVF GILALVTFAI QIISGMVLIL YYKPSIADAF DSATYSIMGE IPFGWLFRH IHATGANFFM AIVYLHMFTG IYYNAYKRPR ELVWIVGWLI YFVLILTALS GYLLPWGQLS YWGFIVTTEI P GSLADAPI LKPIFKAIAE TIVLWMKGGY VVTDVTLGRV FGSHVLIYPL ILLALVGIHL YLVRAAGISN PEGIEYDKKK NP DKFVPFH PYMTLKEGAY VMWYLAVFFF FVFFHISHFL PPENFEPANP LKTPAHIAPE WYLLGYYEVF RSIPSKFWGF VAF NALLLL LLLLPFLDFS PLKSARRRPL FFVMFVIFMI SSMALTILGT MPPTPQNAKL GLIFAALVFA FFISLPIISF IEYG WYKAK GGQQE |
-Macromolecule #3: Cytochrome c
| Macromolecule | Name: Cytochrome c / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Aquifex aeolicus (strain VF5) (bacteria) / Strain: VF5 Aquifex aeolicus (strain VF5) (bacteria) / Strain: VF5 |
| Molecular weight | Theoretical: 27.68017 KDa |
| Sequence | String: MNTWGLIKTI FFAGSTLVFF FLLWFYNPFK HVEHYEVDEE VKAIIDNPWK KTESGKTIAE EGRELFIASC SSCHSLRYDG IYIMSVAAN PKWKNIEKTS GRPVYRFGTL YKDRFFVPKD VYEAFAHDDI QGLKASLGQV PPDLSSMYLA RGEGYLYQFI L NPQKVLPG ...String: MNTWGLIKTI FFAGSTLVFF FLLWFYNPFK HVEHYEVDEE VKAIIDNPWK KTESGKTIAE EGRELFIASC SSCHSLRYDG IYIMSVAAN PKWKNIEKTS GRPVYRFGTL YKDRFFVPKD VYEAFAHDDI QGLKASLGQV PPDLSSMYLA RGEGYLYQFI L NPQKVLPG TTMPQLFNPQ FDPQAKEKVA KIVAYMKSVN TPPPKESAKR TVMGVIVIAY FIVMGLLLWK YRENLLKRLG YH UniProtKB: Cytochrome c |
-Macromolecule #4: FE2/S2 (INORGANIC) CLUSTER
| Macromolecule | Name: FE2/S2 (INORGANIC) CLUSTER / type: ligand / ID: 4 / Number of copies: 2 / Formula: FES |
|---|---|
| Molecular weight | Theoretical: 175.82 Da |
| Chemical component information |  ChemComp-FES: |
-Macromolecule #5: PROTOPORPHYRIN IX CONTAINING FE
| Macromolecule | Name: PROTOPORPHYRIN IX CONTAINING FE / type: ligand / ID: 5 / Number of copies: 4 / Formula: HEM |
|---|---|
| Molecular weight | Theoretical: 616.487 Da |
| Chemical component information |  ChemComp-HEM: |
-Macromolecule #6: 2-[(2~{E},6~{E},10~{Z},14~{Z},18~{Z},23~{R})-3,7,11,15,19,23,27-h...
| Macromolecule | Name: 2-[(2~{E},6~{E},10~{Z},14~{Z},18~{Z},23~{R})-3,7,11,15,19,23,27-heptamethyloctacosa-2,6,10,14,18-pentaenyl]naphthalene-1,4-dione type: ligand / ID: 6 / Number of copies: 6 / Formula: DLX |
|---|---|
| Molecular weight | Theoretical: 639.004 Da |
| Chemical component information | 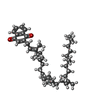 ChemComp-DLX: |
-Macromolecule #7: (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-...
| Macromolecule | Name: (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE type: ligand / ID: 7 / Number of copies: 3 / Formula: PGV |
|---|---|
| Molecular weight | Theoretical: 749.007 Da |
| Chemical component information |  ChemComp-PGV: |
-Macromolecule #8: HEME C
| Macromolecule | Name: HEME C / type: ligand / ID: 8 / Number of copies: 2 / Formula: HEC |
|---|---|
| Molecular weight | Theoretical: 618.503 Da |
| Chemical component information |  ChemComp-HEC: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)