+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0715 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tropomyosin of cardiac thin filament in high-calcium state | |||||||||
 Map data Map data | Tropomyosin in high-calcium state | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cardiac thin filament / CONTRACTILE PROTEIN | |||||||||
| Biological species |  | |||||||||
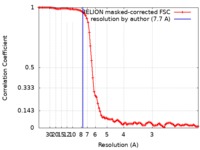
| Method | single particle reconstruction / cryo EM / Resolution: 7.7 Å | |||||||||
 Authors Authors | Oda T / Yanagisawa H | |||||||||
 Citation Citation |  Journal: J Struct Biol / Year: 2020 Journal: J Struct Biol / Year: 2020Title: Cryo-EM structures of cardiac thin filaments reveal the 3D architecture of troponin. Authors: Toshiyuki Oda / Haruaki Yanagisawa / Takeyuki Wakabayashi /  Abstract: Troponin is an essential component of striated muscle and it regulates the sliding of actomyosin system in a calcium-dependent manner. Despite its importance, the structure of troponin has been ...Troponin is an essential component of striated muscle and it regulates the sliding of actomyosin system in a calcium-dependent manner. Despite its importance, the structure of troponin has been elusive due to its high structural heterogeneity. In this study, we analyzed the 3D structures of murine cardiac thin filaments using a cryo-electron microscope equipped with a Volta phase plate (VPP). Contrast enhancement by a VPP enabled us to reconstruct the entire repeat of the thin filament. We determined the orientation of troponin relative to F-actin and tropomyosin, and characterized the interactions between troponin and tropomyosin. This study provides a structural basis for understanding the molecular mechanism of actomyosin system. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0715.map.gz emd_0715.map.gz | 47.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0715-v30.xml emd-0715-v30.xml emd-0715.xml emd-0715.xml | 12 KB 12 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0715_fsc.xml emd_0715_fsc.xml | 9.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_0715.png emd_0715.png | 32.8 KB | ||
| Filedesc metadata |  emd-0715.cif.gz emd-0715.cif.gz | 4.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0715 http://ftp.pdbj.org/pub/emdb/structures/EMD-0715 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0715 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0715 | HTTPS FTP |
-Related structure data
| Related structure data |  6klqMC  0711C  0712C  0714C  0717C  0718C  0796C  0797C  0798C  0799C  0802C  0804C  0805C  0806C  0807C  0808C  6kllC  6klnC  6klpC  6kltC  6kluC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0715.map.gz / Format: CCP4 / Size: 70.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0715.map.gz / Format: CCP4 / Size: 70.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Tropomyosin in high-calcium state | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cardiac thin filament
| Entire | Name: Cardiac thin filament |
|---|---|
| Components |
|
-Supramolecule #1: Cardiac thin filament
| Supramolecule | Name: Cardiac thin filament / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Tropomyosin
| Macromolecule | Name: Tropomyosin / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.507176 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.2 |
| Grid | Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 8460 / Average exposure time: 5.5 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)