[English] 日本語
 Yorodumi
Yorodumi- EMDB-0689: The reconstruction of biotin-bound streptavidin at 3.2 Angstrom r... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0689 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The reconstruction of biotin-bound streptavidin at 3.2 Angstrom resolution | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | streptavidin / CYTOSOLIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Streptomyces avidinii (bacteria) Streptomyces avidinii (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Fan X / Wang J / Lei JL / Wang HW | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Single particle cryo-EM reconstruction of 52 kDa streptavidin at 3.2 Angstrom resolution. Authors: Xiao Fan / Jia Wang / Xing Zhang / Zi Yang / Jin-Can Zhang / Lingyun Zhao / Hai-Lin Peng / Jianlin Lei / Hong-Wei Wang /  Abstract: The fast development of single-particle cryogenic electron microscopy (cryo-EM) has made it more feasible to obtain the 3D structure of well-behaved macromolecules with a molecular weight higher than ...The fast development of single-particle cryogenic electron microscopy (cryo-EM) has made it more feasible to obtain the 3D structure of well-behaved macromolecules with a molecular weight higher than 300 kDa at ~3 Å resolution. However, it remains a challenge to obtain the high-resolution structures of molecules smaller than 200 kDa using single-particle cryo-EM. In this work, we apply the Cs-corrector-VPP-coupled cryo-EM to study the 52 kDa streptavidin (SA) protein supported on a thin layer of graphene and embedded in vitreous ice. We are able to solve both the apo-SA and biotin-bound SA structures at near-atomic resolution using single-particle cryo-EM. We demonstrate that the method has the potential to determine the structures of molecules as small as 39 kDa. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0689.map.gz emd_0689.map.gz | 1.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0689-v30.xml emd-0689-v30.xml emd-0689.xml emd-0689.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0689_fsc.xml emd_0689_fsc.xml | 4.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_0689.png emd_0689.png | 115.1 KB | ||
| Filedesc metadata |  emd-0689.cif.gz emd-0689.cif.gz | 6.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0689 http://ftp.pdbj.org/pub/emdb/structures/EMD-0689 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0689 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0689 | HTTPS FTP |
-Related structure data
| Related structure data |  6j6jMC  0690C  6j6kC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10270 (Title: Single particle reconstruction of 52 kDa biotin-bound state streptavidin at 3.2 Angstrom resolution EMPIAR-10270 (Title: Single particle reconstruction of 52 kDa biotin-bound state streptavidin at 3.2 Angstrom resolutionData size: 5.5 TB Data #1: Uncorrected biotinbound state streptavidin movie stacks, binning 2 from super-resolution stacks. [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0689.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0689.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.053 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Streptavidin with biotin
| Entire | Name: Streptavidin with biotin |
|---|---|
| Components |
|
-Supramolecule #1: Streptavidin with biotin
| Supramolecule | Name: Streptavidin with biotin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Streptomyces avidinii (bacteria) Streptomyces avidinii (bacteria) |
| Molecular weight | Theoretical: 52 KDa |
-Macromolecule #1: Streptavidin
| Macromolecule | Name: Streptavidin / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptomyces avidinii (bacteria) Streptomyces avidinii (bacteria) |
| Molecular weight | Theoretical: 12.596641 KDa |
| Sequence | String: GITGTWYNQL GSTFIVTAGA DGALTGTYES AVGNAESRYV LTGRYDSAPA TDGSGTALGW TVAWKNNYRN AHSATTWSGQ YVGGAEARI NTQWLLTSGT TEANAWKSTL VGHDTFTKVK UniProtKB: Streptavidin |
-Macromolecule #2: BIOTIN
| Macromolecule | Name: BIOTIN / type: ligand / ID: 2 / Number of copies: 4 / Formula: BTN |
|---|---|
| Molecular weight | Theoretical: 244.311 Da |
| Chemical component information | 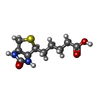 ChemComp-BTN: |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 12 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 285 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE Spherical aberration corrector: spherical aberration corrector |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-32 / Number grids imaged: 1 / Number real images: 1450 / Average exposure time: 2.56 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: -0.8 µm / Nominal defocus min: -0.8 µm / Nominal magnification: 215000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)