+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0552 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | human m-AAA protease AFG3L2, substrate-bound | |||||||||
 Map data Map data | Composite map containing the stitched maps of the ATPase domains from subunits B-E, the ATPase domains from subunits A & F, and the protease domains | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAA+ / ATPase / protease / mitochondria / protein quality control / neurodegeneration / inner membrane / AFG3L2 / m/AAA protease / translocase | |||||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to glutathione / sperm glycocalyx / m-AAA complex / mitochondrial protein processing / regulation of calcium import into the mitochondrion / perinuclear theca / mitochondrial protein quality control / Processing of SMDT1 / mitochondrial calcium ion homeostasis / membrane protein proteolysis ...cellular response to glutathione / sperm glycocalyx / m-AAA complex / mitochondrial protein processing / regulation of calcium import into the mitochondrion / perinuclear theca / mitochondrial protein quality control / Processing of SMDT1 / mitochondrial calcium ion homeostasis / membrane protein proteolysis / calcium import into the mitochondrion / Hydrolases; Acting on acid anhydrides / cristae formation / righting reflex / nerve development / sperm head-tail coupling apparatus / muscle cell development / Hydrolases; Acting on peptide bonds (peptidases); Metalloendopeptidases / mitochondrial fusion / ATP-dependent peptidase activity / neuromuscular junction development / protein autoprocessing / regulation of multicellular organism growth / myelination / Mitochondrial protein degradation / axonogenesis / sperm principal piece / protein maturation / protein catabolic process / protein processing / metalloendopeptidase activity / metallopeptidase activity / unfolded protein binding / sperm midpiece / mitochondrial inner membrane / ATP hydrolysis activity / mitochondrion / proteolysis / zinc ion binding / ATP binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
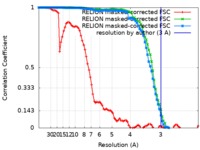
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Lander GC / Puchades C | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2019 Journal: Mol Cell / Year: 2019Title: Unique Structural Features of the Mitochondrial AAA+ Protease AFG3L2 Reveal the Molecular Basis for Activity in Health and Disease. Authors: Cristina Puchades / Bojian Ding / Albert Song / R Luke Wiseman / Gabriel C Lander / Steven E Glynn /  Abstract: Mitochondrial AAA+ quality-control proteases regulate diverse aspects of mitochondrial biology through specialized protein degradation, but the underlying mechanisms of these enzymes remain poorly ...Mitochondrial AAA+ quality-control proteases regulate diverse aspects of mitochondrial biology through specialized protein degradation, but the underlying mechanisms of these enzymes remain poorly defined. The mitochondrial AAA+ protease AFG3L2 is of particular interest, as genetic mutations localized throughout AFG3L2 are linked to diverse neurodegenerative disorders. However, a lack of structural data has limited our understanding of how mutations impact enzymatic function. Here, we used cryoelectron microscopy (cryo-EM) to determine a substrate-bound structure of the catalytic core of human AFG3L2. This structure identifies multiple specialized structural features that integrate with conserved motifs required for ATP-dependent translocation to unfold and degrade targeted proteins. Many disease-relevant mutations localize to these unique structural features of AFG3L2 and distinctly influence its activity and stability. Our results provide a molecular basis for neurological phenotypes associated with different AFG3L2 mutations and establish a structural framework to understand how different members of the AAA+ superfamily achieve specialized biological functions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0552.map.gz emd_0552.map.gz | 5.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0552-v30.xml emd-0552-v30.xml emd-0552.xml emd-0552.xml | 30.8 KB 30.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0552_fsc_1.xml emd_0552_fsc_1.xml emd_0552_fsc_2.xml emd_0552_fsc_2.xml emd_0552_fsc_3.xml emd_0552_fsc_3.xml | 8.6 KB 8.6 KB 8.6 KB | Display Display Display |  FSC data file FSC data file |
| Images |  emd_0552.png emd_0552.png | 331.6 KB | ||
| Filedesc metadata |  emd-0552.cif.gz emd-0552.cif.gz | 7.4 KB | ||
| Others |  emd_0552_additional_1.map.gz emd_0552_additional_1.map.gz emd_0552_additional_2.map.gz emd_0552_additional_2.map.gz emd_0552_additional_3.map.gz emd_0552_additional_3.map.gz emd_0552_additional_4.map.gz emd_0552_additional_4.map.gz emd_0552_half_map_1.map.gz emd_0552_half_map_1.map.gz emd_0552_half_map_2.map.gz emd_0552_half_map_2.map.gz | 7.3 MB 7.3 MB 7.4 MB 7.4 MB 7.3 MB 7.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0552 http://ftp.pdbj.org/pub/emdb/structures/EMD-0552 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0552 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0552 | HTTPS FTP |
-Related structure data
| Related structure data |  6nyyMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0552.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0552.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite map containing the stitched maps of the ATPase domains from subunits B-E, the ATPase domains from subunits A & F, and the protease domains | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: ATPase domains from subunits B-E half map 1
| File | emd_0552_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ATPase domains from subunits B-E half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: ATPase domains from subunits B-E half map 2
| File | emd_0552_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ATPase domains from subunits B-E half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: ATPase domains from subunits A & F half map 2
| File | emd_0552_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ATPase domains from subunits A & F half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: ATPase domains from subunits A & F half map 2
| File | emd_0552_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ATPase domains from subunits A & F half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Protease domains half map 1
| File | emd_0552_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Protease domains half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Protease domains half map 2
| File | emd_0552_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Protease domains half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : AFG3L2
| Entire | Name: AFG3L2 |
|---|---|
| Components |
|
-Supramolecule #1: AFG3L2
| Supramolecule | Name: AFG3L2 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Soluble domains of the AFG3L2, comprising the AAA+ ATPase and protease domains. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 360 KDa |
-Macromolecule #1: AFG3-like protein 2
| Macromolecule | Name: AFG3-like protein 2 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO EC number: Hydrolases; Acting on peptide bonds (peptidases); Metalloendopeptidases |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 58.331629 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNARRGPAGI GRTGRGMGGL FSVGETTAKV LKDEIDVKFK DVAGCEEAKL EIMEFVNFLK NPKQYQDLGA KIPKGAILTG PPGTGKTLL AKATAGEANV PFITVSGSEF LEMFVGVGPA RVRDLFALAR KNAPCILFID QIDAVGRKRG RGNFGGQSEQ E NTLNQLLV ...String: SNARRGPAGI GRTGRGMGGL FSVGETTAKV LKDEIDVKFK DVAGCEEAKL EIMEFVNFLK NPKQYQDLGA KIPKGAILTG PPGTGKTLL AKATAGEANV PFITVSGSEF LEMFVGVGPA RVRDLFALAR KNAPCILFID QIDAVGRKRG RGNFGGQSEQ E NTLNQLLV EMDGFNTTTN VVILAGTNRP DILDPALLRP GRFDRQIFIG PPDIKGRASI FKVHLRPLKL DSTLEKDKLA RK LASLTPG FSGADVANVC NEAALIAARH LSDSINQKHF EQAIERVIGG LEKKTQVLQP EEKKTVAYHQ AGHAVAGWYL EHA DPLLKV SIIPRGKGLG YAQYLPKEQY LYTKEQLLDR MCMTLGGRVS EEIFFGRITT GAQDDLRKVT QSAYAQIVQF GMNE KVGQI SFDLPRQGDM VLEKPYSEAT ARLIDDEVRI LINDAYKRTV ALLTEKKADV EKVALLLLEK EVLDKNDMVE LLGPR PFAE KSTYEEFVEG TGSLDEDTSL PEGLKDWNKE REKEKEEPPG EKVAN UniProtKB: Mitochondrial inner membrane m-AAA protease component AFG3L2 |
-Macromolecule #2: Substrate
| Macromolecule | Name: Substrate / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 799.871 Da |
| Recombinant expression | Organism:  |
| Sequence | String: AAAAAAAAAA A |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 6 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #4: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 4 / Number of copies: 3 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 3 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 1 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
Details: DTT and ATP were added fresh before sample preparation | ||||||||||||||||||
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 6 sec. / Pretreatment - Atmosphere: OTHER Details: Grids were plasma cleaned using a Solarus plasma cleaner (Gatan, Inc.). | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK III Details: Sample was blotted for 3 seconds using Whatman No. 1 filter paper immediately prio to plunge-freezing.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI ARCTICA |
|---|---|
| Temperature | Min: 100.0 K / Max: 100.0 K |
| Alignment procedure | Coma free - Residual tilt: 0.07 mrad |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Frames/image: 1-30 / Number grids imaged: 5 / Number real images: 5707 / Average exposure time: 7.0 sec. / Average electron dose: 40.0 e/Å2 / Details: Data collected with Leginon |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated magnification: 43478 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 18.0 µm / Nominal defocus min: 8.0 µm / Nominal magnification: 36000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | The sequence was threaded onto the PDB ID 6AZ0 and built into the EM density with Coot. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 85 |
| Output model |  PDB-6nyy: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)