+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0476 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of NLRP3 bound to NEK7 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Inflammasome / Activator / Biological process Immunity / Inflammatory response / Innate immunity / Transcription / Transcription regulation Ligand / ATP binding / Nucleotide binding / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationNEK6-subfamily protein kinase / Inhibition of PKR / regulation of NLRP3 inflammasome complex assembly / detection of biotic stimulus / molecular sensor activity / eukaryotic translation initiation factor 2alpha kinase activity / response to interferon-alpha / phosphatidylinositol phosphate binding / negative regulation of osteoblast proliferation / positive regulation of T-helper 2 cell differentiation ...NEK6-subfamily protein kinase / Inhibition of PKR / regulation of NLRP3 inflammasome complex assembly / detection of biotic stimulus / molecular sensor activity / eukaryotic translation initiation factor 2alpha kinase activity / response to interferon-alpha / phosphatidylinositol phosphate binding / negative regulation of osteoblast proliferation / positive regulation of T-helper 2 cell differentiation / Activation of NIMA Kinases NEK9, NEK6, NEK7 / positive regulation of T-helper 2 cell cytokine production / regulation of hematopoietic progenitor cell differentiation / interphase microtubule organizing center / positive regulation of stress-activated MAPK cascade / positive regulation of type 2 immune response / NLRP3 inflammasome complex assembly / NLRP3 inflammasome complex / Nuclear Pore Complex (NPC) Disassembly / protein phosphatase regulator activity / peptidoglycan binding / cysteine-type endopeptidase activator activity / SUMOylation of immune response proteins / osmosensory signaling pathway / regulation of hematopoietic stem cell proliferation / phosphatidylinositol-4-phosphate binding / cellular response to potassium ion / negative regulation of non-canonical NF-kappaB signal transduction / regulation of hematopoietic stem cell differentiation / pattern recognition receptor signaling pathway / regulation of translational initiation / negative regulation of interleukin-1 beta production / negative regulation of viral genome replication / pyroptotic inflammatory response / positive regulation of NLRP3 inflammasome complex assembly / positive regulation of interleukin-4 production / negative regulation of acute inflammatory response / positive regulation of telomere maintenance / microtubule organizing center / The NLRP3 inflammasome / endoplasmic reticulum unfolded protein response / Purinergic signaling in leishmaniasis infection / positive regulation of chemokine production / spindle assembly / signaling adaptor activity / antiviral innate immune response / EML4 and NUDC in mitotic spindle formation / regulation of mitotic cell cycle / molecular function activator activity / cellular response to amino acid starvation / positive regulation of cytokine production / protein maturation / positive regulation of interleukin-1 beta production / non-membrane spanning protein tyrosine kinase activity / non-specific protein-tyrosine kinase / molecular condensate scaffold activity / defense response / positive regulation of non-canonical NF-kappaB signal transduction / Cytoprotection by HMOX1 / : / ADP binding / PKR-mediated signaling / protein homooligomerization / cellular response to virus / Evasion by RSV of host interferon responses / negative regulation of inflammatory response / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / ISG15 antiviral mechanism / response to virus / Metalloprotease DUBs / spindle pole / positive regulation of inflammatory response / SARS-CoV-1 activates/modulates innate immune responses / kinase activity / Interferon alpha/beta signaling / double-stranded RNA binding / protein autophosphorylation / cellular response to lipopolysaccharide / regulation of inflammatory response / protein-macromolecule adaptor activity / defense response to virus / sequence-specific DNA binding / DNA-binding transcription factor binding / molecular adaptor activity / microtubule / protein phosphorylation / protein kinase activity / non-specific serine/threonine protein kinase / positive regulation of MAPK cascade / negative regulation of translation / ribosome / translation / inflammatory response / Golgi membrane / negative regulation of cell population proliferation / innate immune response / protein serine kinase activity / protein serine/threonine kinase activity / apoptotic process / centrosome Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
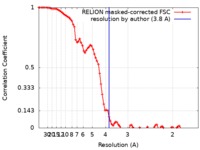
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Sharif H / Wang L / Wu H | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2019 Journal: Nature / Year: 2019Title: Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Authors: Humayun Sharif / Li Wang / Wei Li Wang / Venkat Giri Magupalli / Liudmila Andreeva / Qi Qiao / Arthur V Hauenstein / Zhaolong Wu / Gabriel Núñez / Youdong Mao / Hao Wu /   Abstract: The NLRP3 inflammasome can be activated by stimuli that include nigericin, uric acid crystals, amyloid-β fibrils and extracellular ATP. The mitotic kinase NEK7 licenses the assembly and activation ...The NLRP3 inflammasome can be activated by stimuli that include nigericin, uric acid crystals, amyloid-β fibrils and extracellular ATP. The mitotic kinase NEK7 licenses the assembly and activation of the NLRP3 inflammasome in interphase. Here we report a cryo-electron microscopy structure of inactive human NLRP3 in complex with NEK7, at a resolution of 3.8 Å. The earring-shaped NLRP3 consists of curved leucine-rich-repeat and globular NACHT domains, and the C-terminal lobe of NEK7 nestles against both NLRP3 domains. Structural recognition between NLRP3 and NEK7 is confirmed by mutagenesis both in vitro and in cells. Modelling of an active NLRP3-NEK7 conformation based on the NLRC4 inflammasome predicts an additional contact between an NLRP3-bound NEK7 and a neighbouring NLRP3. Mutations to this interface abolish the ability of NEK7 or NLRP3 to rescue NLRP3 activation in NEK7-knockout or NLRP3-knockout cells. These data suggest that NEK7 bridges adjacent NLRP3 subunits with bipartite interactions to mediate the activation of the NLRP3 inflammasome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0476.map.gz emd_0476.map.gz | 48.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0476-v30.xml emd-0476-v30.xml emd-0476.xml emd-0476.xml | 34.1 KB 34.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0476_fsc.xml emd_0476_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_0476.png emd_0476.png | 150.3 KB | ||
| Filedesc metadata |  emd-0476.cif.gz emd-0476.cif.gz | 9.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0476 http://ftp.pdbj.org/pub/emdb/structures/EMD-0476 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0476 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0476 | HTTPS FTP |
-Related structure data
| Related structure data |  6npyMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0476.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0476.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : NL3-NEK7
| Entire | Name: NL3-NEK7 |
|---|---|
| Components |
|
-Supramolecule #1: NL3-NEK7
| Supramolecule | Name: NL3-NEK7 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|
-Supramolecule #2: NLRP3
| Supramolecule | Name: NLRP3 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #2 Details: The sequence is according to NP_001230062.1 isoform which lacks 2 residues from N-terminus. Total residues are 1034aa as compared to 1036 in UNP Q96P20. There are mutations in YRKKYRKY ...Details: The sequence is according to NP_001230062.1 isoform which lacks 2 residues from N-terminus. Total residues are 1034aa as compared to 1036 in UNP Q96P20. There are mutations in YRKKYRKY instead its IYCAKYRAY mutations were intended to introduce so that the class of MBP and NLRP3 is avoided |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: NEK7
| Supramolecule | Name: NEK7 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Interferon-induced, double-stranded RNA-activated protein kinase,...
| Macromolecule | Name: Interferon-induced, double-stranded RNA-activated protein kinase,Serine/threonine-protein kinase Nek7 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: non-specific serine/threonine protein kinase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 32.01515 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSTDKRFGMD FRIEKKIGRG QFSEVYRAAC RLDGKTVALK KVQIFDLMDA KARADCIKEI DLLAQLNHPN VIKYYVCFIT GNELNIVLE LADAGDLSRM IKHFKKQKRL IPERTVWKYF VQLCSALEHM HSRRVMHRDI KPANVFITAT GVVKLGDLGL G RFFSSKTT ...String: GSTDKRFGMD FRIEKKIGRG QFSEVYRAAC RLDGKTVALK KVQIFDLMDA KARADCIKEI DLLAQLNHPN VIKYYVCFIT GNELNIVLE LADAGDLSRM IKHFKKQKRL IPERTVWKYF VQLCSALEHM HSRRVMHRDI KPANVFITAT GVVKLGDLGL G RFFSSKTT AAHSLVGTPY YMSPERIHEN GYNFKSDIWS LGCLLYEMAA LQSPFYGDKM NLYSLCKKIE QCDYPPLPSD HY SEELRQL VNMCINPDPE KRPDVTYVYD VAKRMHACTA SS UniProtKB: Interferon-induced, double-stranded RNA-activated protein kinase, Serine/threonine-protein kinase Nek7 |
-Macromolecule #2: Isoform 2 of NACHT, LRR and PYD domains-containing protein 3
| Macromolecule | Name: Isoform 2 of NACHT, LRR and PYD domains-containing protein 3 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 117.890984 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASTRCKLAR YLEDLEDVDL KKFKMHLEDY PPQKGCIPLP RGQTEKADHV DLATLMIDFN GEEKAWAMAV WIFAAINRRD LYEKAKRDE PKWGSDNARV SNPTVICQED SIEEEWMGLL EYLSRISICK MKKIYCAKYR AYVRSRFQCI EDRNARLGES V SLNKRYTR ...String: MASTRCKLAR YLEDLEDVDL KKFKMHLEDY PPQKGCIPLP RGQTEKADHV DLATLMIDFN GEEKAWAMAV WIFAAINRRD LYEKAKRDE PKWGSDNARV SNPTVICQED SIEEEWMGLL EYLSRISICK MKKIYCAKYR AYVRSRFQCI EDRNARLGES V SLNKRYTR LRLIKEHRSQ QEREQELLAI GKTKTCESPV SPIKMELLFD PDDEHSEPVH TVVFQGAAGI GKTILARKMM LD WASGTLY QDRFDYLFYI HCREVSLVTQ RSLGDLIMSC CPDPNPPIHK IVRKPSRILF LMDGFDELQG AFDEHIGPLC TDW QKAERG DILLSSLIRK KLLPEASLLI TTRPVALEKL QHLLDHPRHV EILGFSEAKR KEYFFKYFSD EAQARAAFSL IQEN EVLFT MCFIPLVCWI VCTGLKQQME SGKSLAQTSK TTTAVYVFFL SSLLQPRGGS QEHGLCAHLW GLCSLAADGI WNQKI LFEE SDLRNHGLQK ADVSAFLRMN LFQKEVDCEK FYSFIHMTFQ EFFAAMYYLL EEEKEGRTNV PGSRLKLPSR DVTVLL ENY GKFEKGYLIF VVRFLFGLVN QERTSYLEKK LSCKISQQIR LELLKWIEVK AKAKKLQIQP SQLELFYCLY EMQEEDF VQ RAMDYFPKIE INLSTRMDHM VSSFCIENCH RVESLSLGFL HNMPKEEEEE EKEGRHLDMV QCVLPSSSHA ACSHGLVN S HLTSSFCRGL FSVLSTSQSL TELDLSDNSL GDPGMRVLCE TLQHPGCNIR RLWLGRCGLS HECCFDISLV LSSNQKLVE LDLSDNALGD FGIRLLCVGL KHLLCNLKKL WLVSCCLTSA CCQDLASVLS TSHSLTRLYV GENALGDSGV AILCEKAKNP QCNLQKLGL VNSGLTSVCC SALSSVLSTN QNLTHLYLRG NTLGDKGIKL LCEGLLHPDC KLQVLELDNC NLTSHCCWDL S TLLTSSQS LRKLSLGNND LGDLGVMMFC EVLKQQSCLL QNLGLSEMYF NYETKSALET LQEEKPELTV VFEPSW UniProtKB: NACHT, LRR and PYD domains-containing protein 3 |
-Macromolecule #3: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 1 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.1 K / Instrument: FEI VITROBOT MARK II |
- Electron microscopy
Electron microscopy
| Microscope | TFS TITAN THEMIS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -3.0 µm / Nominal defocus min: -1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6npy: |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)