+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0474 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
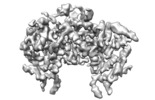
| Title | cryoEM map of NKCC1 | |||||||||
 Map data Map data | cryoEM map of NKCC1 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
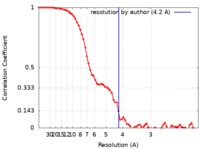
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Benjamin O | |||||||||
 Citation Citation |  Journal: Nature / Year: 2019 Journal: Nature / Year: 2019Title: Structure and mechanism of the cation-chloride cotransporter NKCC1. Authors: Thomas A Chew / Benjamin J Orlando / Jinru Zhang / Naomi R Latorraca / Amy Wang / Scott A Hollingsworth / Dong-Hua Chen / Ron O Dror / Maofu Liao / Liang Feng /  Abstract: Cation-chloride cotransporters (CCCs) mediate the electroneutral transport of chloride, potassium and/or sodium across the membrane. They have critical roles in regulating cell volume, controlling ...Cation-chloride cotransporters (CCCs) mediate the electroneutral transport of chloride, potassium and/or sodium across the membrane. They have critical roles in regulating cell volume, controlling ion absorption and secretion across epithelia, and maintaining intracellular chloride homeostasis. These transporters are primary targets for some of the most commonly prescribed drugs. Here we determined the cryo-electron microscopy structure of the Na-K-Cl cotransporter NKCC1, an extensively studied member of the CCC family, from Danio rerio. The structure defines the architecture of this protein family and reveals how cytosolic and transmembrane domains are strategically positioned for communication. Structural analyses, functional characterizations and computational studies reveal the ion-translocation pathway, ion-binding sites and key residues for transport activity. These results provide insights into ion selectivity, coupling and translocation, and establish a framework for understanding the physiological functions of CCCs and interpreting disease-related mutations. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0474.map.gz emd_0474.map.gz | 24.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0474-v30.xml emd-0474-v30.xml emd-0474.xml emd-0474.xml | 8.9 KB 8.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0474_fsc.xml emd_0474_fsc.xml | 8 KB | Display |  FSC data file FSC data file |
| Images |  emd_0474.png emd_0474.png | 98.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0474 http://ftp.pdbj.org/pub/emdb/structures/EMD-0474 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0474 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0474 | HTTPS FTP |
-Related structure data
| Related structure data |  0470C  0471C  0472C  0473C  0475C  6nphC  6npjC  6npkC  6nplC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0474.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0474.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryoEM map of NKCC1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.055 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : protein sample
| Entire | Name: protein sample |
|---|---|
| Components |
|
-Supramolecule #1: protein sample
| Supramolecule | Name: protein sample / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 7.9 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 96 % / Chamber temperature: 293 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 53.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)