+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0473 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of NKCC1 | |||||||||
 Map data Map data | Membrane protein | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationswim bladder inflation / Cation-coupled Chloride cotransporters / chloride:monoatomic cation symporter activity / sodium:potassium:chloride symporter activity / ammonium transmembrane transport / sodium ion homeostasis / ammonium channel activity / chloride ion homeostasis / ear development / potassium ion homeostasis ...swim bladder inflation / Cation-coupled Chloride cotransporters / chloride:monoatomic cation symporter activity / sodium:potassium:chloride symporter activity / ammonium transmembrane transport / sodium ion homeostasis / ammonium channel activity / chloride ion homeostasis / ear development / potassium ion homeostasis / cell volume homeostasis / inner ear morphogenesis / potassium ion import across plasma membrane / chloride transmembrane transport / sodium ion transmembrane transport / basolateral plasma membrane / apical plasma membrane / metal ion binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Feng L / Liao MF | |||||||||
 Citation Citation |  Journal: Nature / Year: 2019 Journal: Nature / Year: 2019Title: Structure and mechanism of the cation-chloride cotransporter NKCC1. Authors: Thomas A Chew / Benjamin J Orlando / Jinru Zhang / Naomi R Latorraca / Amy Wang / Scott A Hollingsworth / Dong-Hua Chen / Ron O Dror / Maofu Liao / Liang Feng /  Abstract: Cation-chloride cotransporters (CCCs) mediate the electroneutral transport of chloride, potassium and/or sodium across the membrane. They have critical roles in regulating cell volume, controlling ...Cation-chloride cotransporters (CCCs) mediate the electroneutral transport of chloride, potassium and/or sodium across the membrane. They have critical roles in regulating cell volume, controlling ion absorption and secretion across epithelia, and maintaining intracellular chloride homeostasis. These transporters are primary targets for some of the most commonly prescribed drugs. Here we determined the cryo-electron microscopy structure of the Na-K-Cl cotransporter NKCC1, an extensively studied member of the CCC family, from Danio rerio. The structure defines the architecture of this protein family and reveals how cytosolic and transmembrane domains are strategically positioned for communication. Structural analyses, functional characterizations and computational studies reveal the ion-translocation pathway, ion-binding sites and key residues for transport activity. These results provide insights into ion selectivity, coupling and translocation, and establish a framework for understanding the physiological functions of CCCs and interpreting disease-related mutations. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0473.map.gz emd_0473.map.gz | 36.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0473-v30.xml emd-0473-v30.xml emd-0473.xml emd-0473.xml | 13.8 KB 13.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0473.png emd_0473.png | 60.4 KB | ||
| Filedesc metadata |  emd-0473.cif.gz emd-0473.cif.gz | 6.2 KB | ||
| Others |  emd_0473_additional.map.gz emd_0473_additional.map.gz | 36.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0473 http://ftp.pdbj.org/pub/emdb/structures/EMD-0473 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0473 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0473 | HTTPS FTP |
-Related structure data
| Related structure data |  6nplMC  0470C  0471C  0472C  0474C  0475C  6nphC  6npjC  6npkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0473.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0473.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Membrane protein | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.055 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
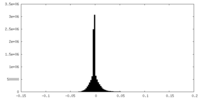
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Membrane protein
| File | emd_0473_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Membrane protein | ||||||||||||
| Projections & Slices |
| ||||||||||||
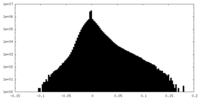
| Density Histograms |
- Sample components
Sample components
-Entire : full length co-transporter
| Entire | Name: full length co-transporter |
|---|---|
| Components |
|
-Supramolecule #1: full length co-transporter
| Supramolecule | Name: full length co-transporter / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Solute carrier family 12 (sodium/potassium/chloride transporter),...
| Macromolecule | Name: Solute carrier family 12 (sodium/potassium/chloride transporter), member 2 type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 100.643445 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KFGWIKGVLV RCMLNIWGVM LFIRMTWIVG QAGIAYSCII VIMATVVTTI TGCSTSAIAT NGFVRGGGAY YLISRSLGPE FGGSIGLIF AFANAVAVAM YVVGFAETVV ELLMDSGLLM IDQTNDIRVI GTITVILLLG ISVAGMEWEA KAQIFLLVIL I TAIFNYFI ...String: KFGWIKGVLV RCMLNIWGVM LFIRMTWIVG QAGIAYSCII VIMATVVTTI TGCSTSAIAT NGFVRGGGAY YLISRSLGPE FGGSIGLIF AFANAVAVAM YVVGFAETVV ELLMDSGLLM IDQTNDIRVI GTITVILLLG ISVAGMEWEA KAQIFLLVIL I TAIFNYFI GSFIAVDSKK KFGFFSYDAG ILAENFGPDF RGQTFFSVFS IFFPAATGIL AGANISGDLA DPQMAIPKGT LL AILITGL VYVGVAISAG ACIVRDATGI ESNFTLISNC TDAACKYGYD FSSCRPTVEG EVSSCKFGLH NDFQVMSVVS GFS PLISAG IFSATLSSAL ASLVSAPKVF QALCKDNIYP GIAIFGKGYG KNNEPLRGYF LTFGIALAFI LIAELNVIAP IISN FFLAS YALINFSVFH ASLANSPGWR PSFKYYNMWA SLAGAILCCV VMFIINWWAA LLTNVIVLSL YIYVSYKKPD VNWGS STQA LTYHQALTHS LQLCGVADHI KTFRPQCLVM TGAPNSRPAI LHLVHAFTKN VGLMLCGHVR ISSRRPNFKE LNSDML RYQ RWLLNNNSKA FYTCVVAEDL RQGTQYMLQA AGLGRLRPNT LVIGFKNDWR IGDIKEVETY INLIHDAFDF QYGVVIL RL REGLDISHIQ GQDDSSGMKD VVVSVDISKD SDGDSSKPSS KATSVQNSPA VQKDKKSPTV PLNVADQRLL DSQQFQQK Q GKGTVDVWWL FDDGGLTLLI PYLIANKKKW KDCKIRVFIG GKINRIDHDR RAMATLLSKF RIDFSDITVL GDINTKPKS EGLTEFAEMI EPYKLREDDM EQEAAEKLKS EEPWRITDNE LELYKAKGNR QIRLNELLKE HSSTANLIVM SMPLARKGAV SSALYMAWL DTLSKDLPPI LLVRGNHQSV LTFYS UniProtKB: Solute carrier family 12 member 2 |
-Macromolecule #2: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Macromolecule #3: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Macromolecule #4: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 4 / Number of copies: 7 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 7.9 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 96 % / Chamber temperature: 293 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 53.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 63659 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)