[English] 日本語
 Yorodumi
Yorodumi- EMDB-0442: Type III-A CRISPR Effector Subcomplex from Staphylococcus epiderm... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0442 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Type III-A CRISPR Effector Subcomplex from Staphylococcus epidermidis RP62a | |||||||||
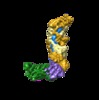
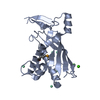
 Map data Map data | Type III-A CRISPR Effector Subcomplex from Staphylococcus epidermidis RP62a, primary map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Staphylococcus epidermidis RP62A (bacteria) Staphylococcus epidermidis RP62A (bacteria) | |||||||||
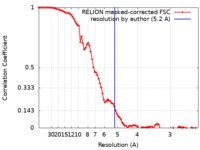
| Method | single particle reconstruction / cryo EM / Resolution: 5.2 Å | |||||||||
 Authors Authors | Dorsey BW / Mondragon A | |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2019 Journal: Nucleic Acids Res / Year: 2019Title: Structural organization of a Type III-A CRISPR effector subcomplex determined by X-ray crystallography and cryo-EM. Authors: Bryan W Dorsey / Lei Huang / Alfonso Mondragón /  Abstract: Clustered regularly interspaced short palindromic repeats (CRISPR) and their associated Cas proteins provide an immune-like response in many prokaryotes against extraneous nucleic acids. CRISPR-Cas ...Clustered regularly interspaced short palindromic repeats (CRISPR) and their associated Cas proteins provide an immune-like response in many prokaryotes against extraneous nucleic acids. CRISPR-Cas systems are classified into different classes and types. Class 1 CRISPR-Cas systems form multi-protein effector complexes that includes a guide RNA (crRNA) used to identify the target for destruction. Here we present crystal structures of Staphylococcus epidermidis Type III-A CRISPR subunits Csm2 and Csm3 and a 5.2 Å resolution single-particle cryo-electron microscopy (cryo-EM) reconstruction of an in vivo assembled effector subcomplex including the crRNA. The structures help to clarify the quaternary architecture of Type III-A effector complexes, and provide details on crRNA binding, target RNA binding and cleavage, and intermolecular interactions essential for effector complex assembly. The structures allow a better understanding of the organization of Type III-A CRISPR effector complexes as well as highlighting the overall similarities and differences with other Class 1 effector complexes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0442.map.gz emd_0442.map.gz | 51.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0442-v30.xml emd-0442-v30.xml emd-0442.xml emd-0442.xml | 13.1 KB 13.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0442_fsc.xml emd_0442_fsc.xml | 8.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_0442.png emd_0442.png | 64.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0442 http://ftp.pdbj.org/pub/emdb/structures/EMD-0442 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0442 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0442 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0442.map.gz / Format: CCP4 / Size: 55.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0442.map.gz / Format: CCP4 / Size: 55.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Type III-A CRISPR Effector Subcomplex from Staphylococcus epidermidis RP62a, primary map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.24 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Type III-A CRISPR Effector Subcomplex from Staphylococcus epiderm...
| Entire | Name: Type III-A CRISPR Effector Subcomplex from Staphylococcus epidermidis RP62a |
|---|---|
| Components |
|
-Supramolecule #1: Type III-A CRISPR Effector Subcomplex from Staphylococcus epiderm...
| Supramolecule | Name: Type III-A CRISPR Effector Subcomplex from Staphylococcus epidermidis RP62a type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Staphylococcus epidermidis RP62A (bacteria) Staphylococcus epidermidis RP62A (bacteria) |
| Molecular weight | Experimental: 255 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.20 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: C-flat-1.2/1.3 4C / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Details: Glow discharged with 15mA. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 99 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: Blot for 4 seconds with +5 force before plunging.. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FS |
|---|---|
| Temperature | Min: 100.0 K / Max: 100.0 K |
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Digitization - Sampling interval: 5.0 µm / Digitization - Frames/image: 1-40 / Number grids imaged: 7 / Number real images: 1037 / Average exposure time: 8.0 sec. / Average electron dose: 37.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated magnification: 40323 / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 2.0 µm / Nominal magnification: 30000 |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)