[English] 日本語
 Yorodumi
Yorodumi- EMDB-0354: CryoEM structure of Leviviridae PP7 coat protein dimer capsid wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0354 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
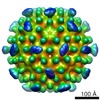
| Title | CryoEM structure of Leviviridae PP7 coat protein dimer capsid with a loop insertion and a C-terminal extension (PP7-aloop-PP7-150loop) | |||||||||
 Map data Map data | PP7-aloop-PP7-150loop T=4 particle | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Pseudomonas phage PP7 (virus) Pseudomonas phage PP7 (virus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.43 Å | |||||||||
 Authors Authors | Liangjun Z / Kopylov M / Carragher B / Potter CS / Finn MG | |||||||||
 Citation Citation |  Journal: ACS Nano / Year: 2019 Journal: ACS Nano / Year: 2019Title: Engineering the PP7 Virus Capsid as a Peptide Display Platform. Authors: Liangjun Zhao / Mykhailo Kopylov / Clinton S Potter / Bridget Carragher / M G Finn /  Abstract: As self-assembling polyvalent nanoscale structures that can tolerate substantial genetic and chemical modification, virus-like particles are useful in a variety of fields. Here we describe the ...As self-assembling polyvalent nanoscale structures that can tolerate substantial genetic and chemical modification, virus-like particles are useful in a variety of fields. Here we describe the genetic modification and structural characterization of the Leviviridae PP7 capsid protein as a platform for the presentation of functional polypeptides. This particle was shown to tolerate the display of sequences from 1 kDa (a cell penetrating peptide) to 14 kDa (the Fc-binding double Z-domain) on its exterior surface as C-terminal genetic fusions to the coat protein. In addition, a dimeric construct allowed the presentation of exogenous loops between capsid monomers and the simultaneous presentation of two different peptides at different positions on the icosahedral structure. The PP7 particle is thereby significantly more tolerant of these types of polypeptide additions than Qβ and MS2, the other Leviviridae-derived VLPs in common use. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0354.map.gz emd_0354.map.gz | 483.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0354-v30.xml emd-0354-v30.xml emd-0354.xml emd-0354.xml | 11.1 KB 11.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_0354.png emd_0354.png | 131.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0354 http://ftp.pdbj.org/pub/emdb/structures/EMD-0354 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0354 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0354 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0354.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0354.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PP7-aloop-PP7-150loop T=4 particle | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.072 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Pseudomonas phage PP7
| Entire | Name:  Pseudomonas phage PP7 (virus) Pseudomonas phage PP7 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Pseudomonas phage PP7
| Supramolecule | Name: Pseudomonas phage PP7 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1 / NCBI-ID: 12023 / Sci species name: Pseudomonas phage PP7 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host system | Organism:  |
| Virus shell | Shell ID: 1 / Name: Icosahedral / Diameter: 340.0 Å / T number (triangulation number): 4 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: PBS |
| Grid | Support film - topology: HOLEY / Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 618 / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 0.002 µm / Nominal defocus min: -0.0007 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















