+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3528 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | nora virus structure | ||||||||||||
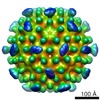
 Map data Map data | nora virus b-factor corrected map. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | nora virus / cryo-em / single particle analysis / capsid / virus | ||||||||||||
| Function / homology | viral capsid / Capsid protein / ORF4 polyprotein Function and homology information Function and homology information | ||||||||||||
| Biological species |  Nora virus Nora virus | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | ||||||||||||
 Authors Authors | Laurinmaki P / Shakeel S | ||||||||||||
| Funding support |  Finland, 3 items Finland, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Rep / Year: 2020 Journal: Sci Rep / Year: 2020Title: Structure of Nora virus at 2.7 Å resolution and implications for receptor binding, capsid stability and taxonomy. Authors: Pasi Laurinmäki / Shabih Shakeel / Jens-Ola Ekström / Pezhman Mohammadi / Dan Hultmark / Sarah J Butcher /    Abstract: Nora virus, a virus of Drosophila, encapsidates one of the largest single-stranded RNA virus genomes known. Its taxonomic affinity is uncertain as it has a picornavirus-like cassette of enzymes for ...Nora virus, a virus of Drosophila, encapsidates one of the largest single-stranded RNA virus genomes known. Its taxonomic affinity is uncertain as it has a picornavirus-like cassette of enzymes for virus replication, but the capsid structure was at the time for genome publication unknown. By solving the structure of the virus, and through sequence comparison, we clear up this taxonomic ambiguity in the invertebrate RNA virosphere. Despite the lack of detectable similarity in the amino acid sequences, the 2.7 Å resolution cryoEM map showed Nora virus to have T = 1 symmetry with the characteristic capsid protein β-barrels found in all the viruses in the Picornavirales order. Strikingly, α-helical bundles formed from the extended C-termini of capsid protein VP4B and VP4C protrude from the capsid surface. They are similar to signalling molecule folds and implicated in virus entry. Unlike other viruses of Picornavirales, no intra-pentamer stabilizing annulus was seen, instead the intra-pentamer stability comes from the interaction of VP4C and VP4B N-termini. Finally, intertwining of the N-termini of two-fold symmetry-related VP4A capsid proteins and RNA, provides inter-pentamer stability. Based on its distinct structural elements and the genetic distance to other picorna-like viruses we propose that Nora virus, and a small group of related viruses, should have its own family within the order Picornavirales. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3528.map.gz emd_3528.map.gz | 380.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3528-v30.xml emd-3528-v30.xml emd-3528.xml emd-3528.xml | 22.2 KB 22.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3528_fsc.xml emd_3528_fsc.xml | 16.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_3528.png emd_3528.png | 68.3 KB | ||
| Masks |  emd_3528_msk_1.map emd_3528_msk_1.map | 421.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-3528.cif.gz emd-3528.cif.gz | 6.5 KB | ||
| Others |  emd_3528_additional.map.gz emd_3528_additional.map.gz emd_3528_additional_1.map.gz emd_3528_additional_1.map.gz emd_3528_half_map_1.map.gz emd_3528_half_map_1.map.gz emd_3528_half_map_2.map.gz emd_3528_half_map_2.map.gz | 334.6 MB 334.6 MB 336.4 MB 336.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3528 http://ftp.pdbj.org/pub/emdb/structures/EMD-3528 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3528 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3528 | HTTPS FTP |
-Related structure data
| Related structure data |  5mm2MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10088 (Title: Single particle dataset of Nora virus / Data size: 3.6 TB EMPIAR-10088 (Title: Single particle dataset of Nora virus / Data size: 3.6 TBData #1: Unaligned multi-frame micrographs of Nora virus [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3528.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3528.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | nora virus b-factor corrected map. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_3528_msk_1.map emd_3528_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: nora virus, not b-factor corrected
| File | emd_3528_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | nora virus, not b-factor corrected | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: nora virus, not b-factor corrected
| File | emd_3528_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | nora virus, not b-factor corrected | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_3528_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_3528_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Nora virus
| Entire | Name:  Nora virus Nora virus |
|---|---|
| Components |
|
-Supramolecule #1: Nora virus
| Supramolecule | Name: Nora virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Drosophila Nora virus strain Umea 2007 was derived from an infectious cDNA clone (accession GQ257737). NCBI-ID: 363716 / Sci species name: Nora virus / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
| Virus shell | Shell ID: 1 / T number (triangulation number): 1 |
-Macromolecule #1: capsid protein VP4C
| Macromolecule | Name: capsid protein VP4C / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Nora virus Nora virus |
| Molecular weight | Theoretical: 45.258152 KDa |
| Sequence | String: SLPENAPNAV SNPQQFITPA TALSAEEYNV HEALGETEEL ELDEFPVLVF KGNVPVDSVT SIPLDLATIY DFAWDGEQNA ISQKFQRFA HLIPKSAGGF GPVIGNYTIT ANLPTGVAGR ILHNCLPGDC VDLAVSRIFG LKSLLGVAGT AVSAIGGPLL N GLVNTAAP ...String: SLPENAPNAV SNPQQFITPA TALSAEEYNV HEALGETEEL ELDEFPVLVF KGNVPVDSVT SIPLDLATIY DFAWDGEQNA ISQKFQRFA HLIPKSAGGF GPVIGNYTIT ANLPTGVAGR ILHNCLPGDC VDLAVSRIFG LKSLLGVAGT AVSAIGGPLL N GLVNTAAP ILSGAAHAIG GNVVGGLADA VIDIGSNLLT PKEKEQPSAN SSAISGDIPI SRFVEMLKYV KENYQDNPVF PT LLVEPQN FISNAMTALK TIPIEVFANM RNVKVERNLF DRTVVPTVKE ATLADIVIPN HMYGYILRDF LQNKRAFQSG TKQ NVYFQQ FLTVLSQRNI RTHITLNDIT SCSIDSESIA NKIERVKHYL STNSSGETTE EFSRTDTGLL PITTRKIVLG ESKR RTERY VAETVFPSVR Q UniProtKB: ORF4 polyprotein |
-Macromolecule #2: capsid protein VP4B
| Macromolecule | Name: capsid protein VP4B / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Nora virus Nora virus |
| Molecular weight | Theoretical: 28.166934 KDa |
| Sequence | String: ADNEVTAEGG KLVQELVYDH SAIPVAPVVE TQAEQPEVPV SLVATRKNDT GHLATKWYDF AKISLSNPAN MNWTTLTIDP YNNVTLSRD GESMVLPWRR NVWTTGSKSI GYIRTMVAQI NIPRPPQISG VLEVKDSINN SSISLVEFGG KVEIPIIPKV M NGLATTAS ...String: ADNEVTAEGG KLVQELVYDH SAIPVAPVVE TQAEQPEVPV SLVATRKNDT GHLATKWYDF AKISLSNPAN MNWTTLTIDP YNNVTLSRD GESMVLPWRR NVWTTGSKSI GYIRTMVAQI NIPRPPQISG VLEVKDSINN SSISLVEFGG KVEIPIIPKV M NGLATTAS LPRHRLNPWM RTAESKVELQ YRIIAFNRTS DIADLNVSVL LRPGDSQFQL PMKPDNNVDT RHFELVEALM YH YDSLRIR GEEQ UniProtKB: ORF4 polyprotein |
-Macromolecule #3: Capsid protein VP4A
| Macromolecule | Name: Capsid protein VP4A / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Nora virus Nora virus |
| Molecular weight | Theoretical: 29.019826 KDa |
| Sequence | String: MQNPTQTMHI YDMPLRVIAG LSTLAKTTEE DDNTSTGIVV SEVGEPQVVN HPAWIDPFVA YQLRAPRKNI TPDFIFGRAD IGNAFSAFL PRRFSAPAVG TRLVVDPVFT YQQRTVLGLY NYFHADFYYI VHVPAPLGTG IYLKIYAPEF DTTTVTRGIR F KPSASPTI ...String: MQNPTQTMHI YDMPLRVIAG LSTLAKTTEE DDNTSTGIVV SEVGEPQVVN HPAWIDPFVA YQLRAPRKNI TPDFIFGRAD IGNAFSAFL PRRFSAPAVG TRLVVDPVFT YQQRTVLGLY NYFHADFYYI VHVPAPLGTG IYLKIYAPEF DTTTVTRGIR F KPSASPTI ALSVPWSNDL STVETSVGRV GQSGGSIVIE TIEDNSNETV NTPLSITVWC CMANIKATGY RHADTSAYNE KG MNFIPVP VPKPPVPPTK PITGEEQ UniProtKB: Capsid protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 / Details: 10mM Tris-HCl pH 7.4. |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 75 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 295 K / Instrument: LEICA EM GP |
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 3516 / Average exposure time: 8.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 47170 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Target criteria: Cross-correlation coefficient |
|---|---|
| Output model |  PDB-5mm2: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






























































