[English] 日本語
 Yorodumi
Yorodumi- EMDB-0274: Cryo-EM structure of cardiac amyloid fibrils from an immunoglobul... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0274 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of cardiac amyloid fibrils from an immunoglobulin light chain (AL) amyloidosis patient. | |||||||||||||||
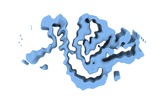
 Map data Map data | 3D helical reconstrutcion of an amyloid fibrils from an immunoglobulin light chain (AL) amyloidosis patient. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | immunoglobulin light chains / amyloid cardiomyopathy / ex-vivo cryo-EM / helical reconstruction / PROTEIN FIBRIL | |||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||
| Method | helical reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||||||||
 Authors Authors | Swuec P | |||||||||||||||
| Funding support |  Italy, 4 items Italy, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2019 Journal: Nat Commun / Year: 2019Title: Cryo-EM structure of cardiac amyloid fibrils from an immunoglobulin light chain AL amyloidosis patient. Authors: Paolo Swuec / Francesca Lavatelli / Masayoshi Tasaki / Cristina Paissoni / Paola Rognoni / Martina Maritan / Francesca Brambilla / Paolo Milani / Pierluigi Mauri / Carlo Camilloni / Giovanni ...Authors: Paolo Swuec / Francesca Lavatelli / Masayoshi Tasaki / Cristina Paissoni / Paola Rognoni / Martina Maritan / Francesca Brambilla / Paolo Milani / Pierluigi Mauri / Carlo Camilloni / Giovanni Palladini / Giampaolo Merlini / Stefano Ricagno / Martino Bolognesi /   Abstract: Systemic light chain amyloidosis (AL) is a life-threatening disease caused by aggregation and deposition of monoclonal immunoglobulin light chains (LC) in target organs. Severity of heart ...Systemic light chain amyloidosis (AL) is a life-threatening disease caused by aggregation and deposition of monoclonal immunoglobulin light chains (LC) in target organs. Severity of heart involvement is the most important factor determining prognosis. Here, we report the 4.0 Å resolution cryo-electron microscopy map and molecular model of amyloid fibrils extracted from the heart of an AL amyloidosis patient with severe amyloid cardiomyopathy. The helical fibrils are composed of a single protofilament, showing typical 4.9 Å stacking and cross-β architecture. Two distinct polypeptide stretches (total of 77 residues) from the LC variable domain (V) fit the fibril density. Despite V high sequence variability, residues stabilizing the fibril core are conserved through different cardiotoxic V, highlighting structural motifs that may be common to misfolding-prone LCs. Our data shed light on the architecture of LC amyloids, correlate amino acid sequences with fibril assembly, providing the grounds for development of innovative medicines. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0274.map.gz emd_0274.map.gz | 4.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0274-v30.xml emd-0274-v30.xml emd-0274.xml emd-0274.xml | 12.1 KB 12.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0274_fsc.xml emd_0274_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_0274.png emd_0274.png | 121.4 KB | ||
| Filedesc metadata |  emd-0274.cif.gz emd-0274.cif.gz | 5.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0274 http://ftp.pdbj.org/pub/emdb/structures/EMD-0274 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0274 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0274 | HTTPS FTP |
-Related structure data
| Related structure data |  6hudMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0274.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0274.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D helical reconstrutcion of an amyloid fibrils from an immunoglobulin light chain (AL) amyloidosis patient. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.887 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cardiac amyloid fibril from an immunoglobulin light chain (AL) am...
| Entire | Name: Cardiac amyloid fibril from an immunoglobulin light chain (AL) amyloidosis patient |
|---|---|
| Components |
|
-Supramolecule #1: Cardiac amyloid fibril from an immunoglobulin light chain (AL) am...
| Supramolecule | Name: Cardiac amyloid fibril from an immunoglobulin light chain (AL) amyloidosis patient type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Tissue: Heart Homo sapiens (human) / Tissue: Heart |
-Macromolecule #1: Monoclonal immunoglobulin light chains (LC)
| Macromolecule | Name: Monoclonal immunoglobulin light chains (LC) / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / Tissue: Heart Homo sapiens (human) / Tissue: Heart |
| Molecular weight | Theoretical: 14.620864 KDa |
| Sequence | String: NFMLTQPHSV SESPGKTLTI SCTGSSASIA SHYVQWYQQR PGGAPTTLIY ENDQRPSEVP DRFSGSIDSS SNSASLTISG LKTEDEADY YCQSYDGNNH WVFGGGTKLT VLSQPKAAPS VTLFPPSSEE LQANKAT |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.01 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number real images: 1680 / Average exposure time: 1.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 120000 |
| Sample stage | Specimen holder model: OTHER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-6hud: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















