+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0179 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM map of the Fatty Acid Synthase from S. cerevisiae | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 24.6 Å | |||||||||
 Authors Authors | D'Imprima E / Floris D / Sanchez R / Joppe M / Grininger M / Kuehlbrandt W | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: Protein denaturation at the air-water interface and how to prevent it. Authors: Edoardo D'Imprima / Davide Floris / Mirko Joppe / Ricardo Sánchez / Martin Grininger / Werner Kühlbrandt /  Abstract: Electron cryo-microscopy analyzes the structure of proteins and protein complexes in vitrified solution. Proteins tend to adsorb to the air-water interface in unsupported films of aqueous solution, ...Electron cryo-microscopy analyzes the structure of proteins and protein complexes in vitrified solution. Proteins tend to adsorb to the air-water interface in unsupported films of aqueous solution, which can result in partial or complete denaturation. We investigated the structure of yeast fatty acid synthase at the air-water interface by electron cryo-tomography and single-particle image processing. Around 90% of complexes adsorbed to the air-water interface are partly denatured. We show that the unfolded regions face the air-water interface. Denaturation by contact with air may happen at any stage of specimen preparation. Denaturation at the air-water interface is completely avoided when the complex is plunge-frozen on a substrate of hydrophilized graphene. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0179.map.gz emd_0179.map.gz | 9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0179-v30.xml emd-0179-v30.xml emd-0179.xml emd-0179.xml | 8.2 KB 8.2 KB | Display Display |  EMDB header EMDB header |
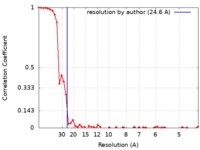
| FSC (resolution estimation) |  emd_0179_fsc.xml emd_0179_fsc.xml | 6 KB | Display |  FSC data file FSC data file |
| Images |  emd_0179.png emd_0179.png | 76 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0179 http://ftp.pdbj.org/pub/emdb/structures/EMD-0179 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0179 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0179 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0179.map.gz / Format: CCP4 / Size: 10.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0179.map.gz / Format: CCP4 / Size: 10.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cryo-EM map of the Fatty Acid Synthase from S. cerevisiae
| Entire | Name: Cryo-EM map of the Fatty Acid Synthase from S. cerevisiae |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM map of the Fatty Acid Synthase from S. cerevisiae
| Supramolecule | Name: Cryo-EM map of the Fatty Acid Synthase from S. cerevisiae type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 2.6 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 6.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 2.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)