[English] 日本語
 Yorodumi
Yorodumi- EMDB-0172: Cryo-EM structure of the archaeal extremophilic internal membrane... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0172 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
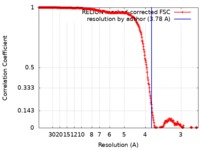
| Title | Cryo-EM structure of the archaeal extremophilic internal membrane containing Haloarcula hispanica icosahedral virus 2 (HHIV-2) at 3.78 Angstroms resolution. | |||||||||||||||
 Map data Map data | Map derived from RELION postprocessed map. The map was then centred at 0,0,0 and finally used for model refinement. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | single vertical beta-barrel virus / archaeal / membrane-containing / quasi-atomic resolution / VIRUS | |||||||||||||||
| Function / homology | VP7 / VP4 / Uncharacterized protein Function and homology information Function and homology information | |||||||||||||||
| Biological species |  Haloarcula hispanica icosahedral virus 2 Haloarcula hispanica icosahedral virus 2 | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.78 Å | |||||||||||||||
 Authors Authors | Abrescia NG / Santos-Perez I | |||||||||||||||
| Funding support |  Spain, Spain,  Finland, 4 items Finland, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Structure / Year: 2015 Journal: Structure / Year: 2015Title: Insight into the Assembly of Viruses with Vertical Single β-barrel Major Capsid Proteins. Authors: David Gil-Carton / Salla T Jaakkola / Diego Charro / Bibiana Peralta / Daniel Castaño-Díez / Hanna M Oksanen / Dennis H Bamford / Nicola G A Abrescia /    Abstract: Archaeal viruses constitute the least explored niche within the virosphere. Structure-based approaches have revealed close relationships between viruses infecting organisms from different domains of ...Archaeal viruses constitute the least explored niche within the virosphere. Structure-based approaches have revealed close relationships between viruses infecting organisms from different domains of life. Here, using biochemical and cryo-electron microscopy techniques, we solved the structure of euryarchaeal, halophilic, internal membrane-containing Haloarcula hispanica icosahedral virus 2 (HHIV-2). We show that the density of the two major capsid proteins (MCPs) recapitulates vertical single β-barrel proteins and that disulfide bridges stabilize the capsid. Below, ordered density is visible close to the membrane and at the five-fold vertices underneath the host-interacting vertex complex underpinning membrane-protein interactions. The HHIV-2 structure exemplifies the division of conserved architectural elements of a virion, such as the capsid, from those that evolve rapidly due to selective environmental pressure such as host-recognizing structures. We propose that in viruses with two vertical single β-barrel MCPs the vesicle is indispensable, and membrane-protein interactions serve as protein-railings for guiding the assembly. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0172.map.gz emd_0172.map.gz | 1.5 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0172-v30.xml emd-0172-v30.xml emd-0172.xml emd-0172.xml | 29.9 KB 29.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0172_fsc.xml emd_0172_fsc.xml | 26.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_0172.png emd_0172.png | 219.2 KB | ||
| Masks |  emd_0172_msk_1.map emd_0172_msk_1.map | 1.6 GB |  Mask map Mask map | |
| Filedesc metadata |  emd-0172.cif.gz emd-0172.cif.gz | 6.8 KB | ||
| Others |  emd_0172_additional.map.gz emd_0172_additional.map.gz emd_0172_half_map_1.map.gz emd_0172_half_map_1.map.gz emd_0172_half_map_2.map.gz emd_0172_half_map_2.map.gz | 1.5 GB 1.3 GB 1.3 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0172 http://ftp.pdbj.org/pub/emdb/structures/EMD-0172 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0172 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0172 | HTTPS FTP |
-Related structure data
| Related structure data |  6h82MC  0050C  0072C  0073C  0131C  0174C  6h9cC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0172.map.gz / Format: CCP4 / Size: 1.6 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0172.map.gz / Format: CCP4 / Size: 1.6 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map derived from RELION postprocessed map. The map was then centred at 0,0,0 and finally used for model refinement. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_0172_msk_1.map emd_0172_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
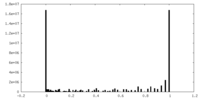
| Density Histograms |
-Additional map: Relion post-processed map bfactor -40
| File | emd_0172_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Relion post-processed map bfactor -40 | ||||||||||||
| Projections & Slices |
| ||||||||||||
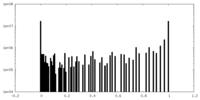
| Density Histograms |
-Half map: Half maps for gold-standard FSC and generation of...
| File | emd_0172_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half maps for gold-standard FSC and generation of both submitted postprocessed maps. | ||||||||||||
| Projections & Slices |
| ||||||||||||
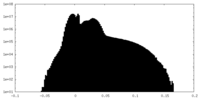
| Density Histograms |
-Half map: #1
| File | emd_0172_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
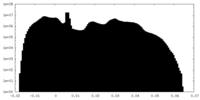
| Density Histograms |
- Sample components
Sample components
-Entire : Haloarcula hispanica icosahedral virus 2
| Entire | Name:  Haloarcula hispanica icosahedral virus 2 Haloarcula hispanica icosahedral virus 2 |
|---|---|
| Components |
|
-Supramolecule #1: Haloarcula hispanica icosahedral virus 2
| Supramolecule | Name: Haloarcula hispanica icosahedral virus 2 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 1154689 / Sci species name: Haloarcula hispanica icosahedral virus 2 / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Haloarcula hispanica ATCC 33960 (Halophile) Haloarcula hispanica ATCC 33960 (Halophile) |
-Macromolecule #1: VP4
| Macromolecule | Name: VP4 / type: protein_or_peptide / ID: 1 / Details: http://mit.cicbiogune.int:39000/projects/P2/W1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Haloarcula hispanica icosahedral virus 2 Haloarcula hispanica icosahedral virus 2 |
| Molecular weight | Theoretical: 25.585746 KDa |
| Sequence | String: QTQEYTINHT GGVLGDSYVT TASNQTSPQR ETAVLSFECP RKFEEINYVG QRDATRFVPR TTESITGSAN DDTVVDLTAN IQPVAGEEV IAEQDYPVAV AYNVTQGVEV DVVDADYAAD TVTLGTNPAD GDEVKVWPIM SDGDVQFRLI NQFGQEEGRV Y PWSTPLYR ...String: QTQEYTINHT GGVLGDSYVT TASNQTSPQR ETAVLSFECP RKFEEINYVG QRDATRFVPR TTESITGSAN DDTVVDLTAN IQPVAGEEV IAEQDYPVAV AYNVTQGVEV DVVDADYAAD TVTLGTNPAD GDEVKVWPIM SDGDVQFRLI NQFGQEEGRV Y PWSTPLYR WHDFPQLKRG REINLHGSAS WSENETLEIL LDAPQALTWE DSDYPRGQYV TTLEQDVEIT L UniProtKB: VP4 |
-Macromolecule #2: VP7
| Macromolecule | Name: VP7 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Haloarcula hispanica icosahedral virus 2 Haloarcula hispanica icosahedral virus 2 |
| Molecular weight | Theoretical: 18.473355 KDa |
| Sequence | String: PEIGNNGAEK QISLHKGQPF IDTQDVGAAD PNTPAVTIEG PSDYVIAIDA GTPVAPEFRD ANGDKLDPST RVTIQKCDKQ GNPLGDGIV FSDTLGRFEY SKMRSDPDYM RKTTTSLMID EREIVKIFVE VPPNANGMDA DNSRITIGDD TSDYGKAVGI V EHGDLSPA ESKA UniProtKB: VP7 |
-Macromolecule #3: VP7
| Macromolecule | Name: VP7 / type: protein_or_peptide / ID: 3 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Haloarcula hispanica icosahedral virus 2 Haloarcula hispanica icosahedral virus 2 |
| Molecular weight | Theoretical: 18.857811 KDa |
| Sequence | String: PEIGNNGAEK QISLHKGQPF IDTQDVGAAD PNTPAVTIEG PSDYVIAIDA GTPVAPEFRD ANGDKLDPST RVTIQKCDKQ GNPLGDGIV FSDTLGRFEY SKMRSDPDYM RKTTTSLMID EREIVKIFVE VPPNANGMDA DNSRITIGDD TSDYGKAVGI V EHGDLSPA ESKAVRQ UniProtKB: VP7 |
-Macromolecule #4: VP7
| Macromolecule | Name: VP7 / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Haloarcula hispanica icosahedral virus 2 Haloarcula hispanica icosahedral virus 2 |
| Molecular weight | Theoretical: 17.347154 KDa |
| Sequence | String: IGNNGAEKQI SLHKGQPFID TQDVGAADPN TPAVTIEGPS DYVIAIDAGT PVAPEFRDAN GDKLDPSTRV TIQKCDKQGN PLGDGIVFS DTLGRFEYSK MRSDPDYMRK TTTSLMIDER EIVKIFVEVP PNANGMDADN SRITIGDDTS DYGKAVGIVE H G UniProtKB: VP7 |
-Macromolecule #5: Uncharacterized protein
| Macromolecule | Name: Uncharacterized protein / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Haloarcula hispanica icosahedral virus 2 Haloarcula hispanica icosahedral virus 2 |
| Molecular weight | Theoretical: 14.206529 KDa |
| Sequence | String: QTADGRVGLV PVNSYVTLET DDLDTDEHPV TDAGTVALEP GESAPIVRYD LGQPAAVYAV GATDEANVEY ELKVNNSKTV GGRTNSPLG VLNTPFSFVE KLGGAIPCET AATYWAHYSS DATGTVELAG RMHIEV UniProtKB: Uncharacterized protein |
-Macromolecule #6: VP16 (vertex complex)
| Macromolecule | Name: VP16 (vertex complex) / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Haloarcula hispanica icosahedral virus 2 Haloarcula hispanica icosahedral virus 2 |
| Molecular weight | Theoretical: 18.485723 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Macromolecule #7: GPS III
| Macromolecule | Name: GPS III / type: protein_or_peptide / ID: 7 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Haloarcula hispanica icosahedral virus 2 Haloarcula hispanica icosahedral virus 2 |
| Molecular weight | Theoretical: 8.954028 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) |
-Macromolecule #8: polypeptide stretch (vertex complex)
| Macromolecule | Name: polypeptide stretch (vertex complex) / type: protein_or_peptide / ID: 8 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Haloarcula hispanica icosahedral virus 2 Haloarcula hispanica icosahedral virus 2 |
| Molecular weight | Theoretical: 2.145636 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.2 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 | |||||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Y (Sec.)
Y (Sec.) X (Row.)
X (Row.) Z (Col.)
Z (Col.)