+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Template-free detection and classification of microsomal membrane bound complexes | |||||||||
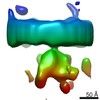
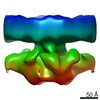
 マップデータ マップデータ | Translocon complex associated with ribosome | |||||||||
 試料 試料 |
| |||||||||
| 生物種 |  | |||||||||
| 手法 | サブトモグラム平均法 / クライオ電子顕微鏡法 / 解像度: 22.0 Å | |||||||||
 データ登録者 データ登録者 | Martinez-Sanchez A / Lucic V | |||||||||
 引用 引用 |  ジャーナル: Nat Commun / 年: 2015 ジャーナル: Nat Commun / 年: 2015タイトル: Structure of the native Sec61 protein-conducting channel. 著者: Stefan Pfeffer / Laura Burbaum / Pia Unverdorben / Markus Pech / Yuxiang Chen / Richard Zimmermann / Roland Beckmann / Friedrich Förster /  要旨: In mammalian cells, secretory and membrane proteins are translocated across or inserted into the endoplasmic reticulum (ER) membrane by the universally conserved protein-conducting channel Sec61, ...In mammalian cells, secretory and membrane proteins are translocated across or inserted into the endoplasmic reticulum (ER) membrane by the universally conserved protein-conducting channel Sec61, which has been structurally studied in isolated, detergent-solubilized states. Here we structurally and functionally characterize native, non-solubilized ribosome-Sec61 complexes on rough ER vesicles using cryo-electron tomography and ribosome profiling. Surprisingly, the 9-Å resolution subtomogram average reveals Sec61 in a laterally open conformation, even though the channel is not in the process of inserting membrane proteins into the lipid bilayer. In contrast to recent mechanistic models for polypeptide translocation and insertion, our results indicate that the laterally open conformation of Sec61 is the only conformation present in the ribosome-bound translocon complex, independent of its functional state. Consistent with earlier functional studies, our structure suggests that the ribosome alone, even without a nascent chain, is sufficient for lateral opening of Sec61 in a lipid environment. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|---|
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_0085.map.gz emd_0085.map.gz | 14.5 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-0085-v30.xml emd-0085-v30.xml emd-0085.xml emd-0085.xml | 17.1 KB 17.1 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_0085_fsc.xml emd_0085_fsc.xml | 5.8 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_0085.png emd_0085.png | 38.1 KB | ||
| その他 |  emd_0085_half_map_1.map.gz emd_0085_half_map_1.map.gz emd_0085_half_map_2.map.gz emd_0085_half_map_2.map.gz | 11.9 MB 11.9 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0085 http://ftp.pdbj.org/pub/emdb/structures/EMD-0085 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0085 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0085 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_0085_validation.pdf.gz emd_0085_validation.pdf.gz | 456 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_0085_full_validation.pdf.gz emd_0085_full_validation.pdf.gz | 455.1 KB | 表示 | |
| XML形式データ |  emd_0085_validation.xml.gz emd_0085_validation.xml.gz | 11.6 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0085 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0085 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0085 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-0085 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_0085.map.gz / 形式: CCP4 / 大きさ: 15.6 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_0085.map.gz / 形式: CCP4 / 大きさ: 15.6 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Translocon complex associated with ribosome | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 2.62 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-ハーフマップ: Translocon complex associated with ribosome, 1st half
| ファイル | emd_0085_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Translocon complex associated with ribosome, 1st half | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: Translocon complex associated with ribosome, 2nd half
| ファイル | emd_0085_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Translocon complex associated with ribosome, 2nd half | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Mammalian ribosome bound to the native protein translocon on cani...
| 全体 | 名称: Mammalian ribosome bound to the native protein translocon on canine pancreatic ER vesicles |
|---|---|
| 要素 |
|
-超分子 #1: Mammalian ribosome bound to the native protein translocon on cani...
| 超分子 | 名称: Mammalian ribosome bound to the native protein translocon on canine pancreatic ER vesicles タイプ: complex / ID: 1 / 親要素: 0 / 詳細: The same sample as the one used for EMD-3068 - 72 |
|---|---|
| 由来(天然) | 生物種:  |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | サブトモグラム平均法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 2.0 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 緩衝液 | pH: 7.6 構成要素:
詳細: The same as for EMD-3068 - 72 | ||||||||||||
| 凍結 | 凍結剤: ETHANE-PROPANE / チャンバー内湿度: 70 % / 装置: FEI VITROBOT MARK IV / 詳細: Blot 3 seconds before plunging.. | ||||||||||||
| 詳細 | The same sample was used for EMD-3068 - 72 |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 詳細 | The same as for EMD-3068 - 72 |
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: COUNTING / 平均電子線量: 0.75 e/Å2 / 詳細: The same as for EMD-3068 - 72 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 4.0 µm / 最小 デフォーカス(公称値): 3.0 µm |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 精密化 | プロトコル: AB INITIO MODEL |
|---|
 ムービー
ムービー コントローラー
コントローラー























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)