[English] 日本語
 Yorodumi
Yorodumi- SASDFL8: GON7, the fifth subunit of human KEOPS, bound to LAGE3-OSGEP (EKC... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: SASBDB / ID: SASDFL8 |
|---|---|
 Sample Sample | GON7, the fifth subunit of human KEOPS, bound to LAGE3-OSGEP (EKC/KEOPS complex subunit:Probable tRNA N6-adenosine threonylcarbamoyltransferase)
|
| Function / homology |  Function and homology information Function and homology informationN6-L-threonylcarbamoyladenine synthase / tRNA N(6)-L-threonylcarbamoyladenine synthase activity / EKC/KEOPS complex / tRNA threonylcarbamoyladenosine modification / tRNA modification in the nucleus and cytosol / tRNA modification / nucleolus / nucleoplasm / metal ion binding / nucleus ...N6-L-threonylcarbamoyladenine synthase / tRNA N(6)-L-threonylcarbamoyladenine synthase activity / EKC/KEOPS complex / tRNA threonylcarbamoyladenosine modification / tRNA modification in the nucleus and cytosol / tRNA modification / nucleolus / nucleoplasm / metal ion binding / nucleus / cytosol / cytoplasm Similarity search - Function |
| Biological species |  Homo sapiens (human) Homo sapiens (human) |
 Citation Citation |  Date: 2019 Dec Date: 2019 DecTitle: Defects in t6A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome Authors: Arrondel C / Missoury S / Snoek R / Patat J / Menara G / Collinet B / Liger D / Durand D / Gribouval O / Boyer O / Buscara L / Martin G / Machuca E / Nevo F / Lescop E / Braun D / Boschat A ...Authors: Arrondel C / Missoury S / Snoek R / Patat J / Menara G / Collinet B / Liger D / Durand D / Gribouval O / Boyer O / Buscara L / Martin G / Machuca E / Nevo F / Lescop E / Braun D / Boschat A / Sanquer S / Guerrera I / Revy P / Parisot M / Masson C / Boddaert N / Charbit M / Decramer S / Novo R / Macher M / Ranchin B / Bacchetta J / Laurent A / Collardeau-Frachon S / van Eerde A / Hildebrandt F / Magen D / Antignac C / van Tilbeurgh H |
 Contact author Contact author |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
-Models
| Model #3207 |  Type: atomic / Chi-square value: 0.308 / P-value: 0.001285  Search similar-shape structures of this assembly by Omokage search (details) Search similar-shape structures of this assembly by Omokage search (details) |
|---|
- Sample
Sample
 Sample Sample | Name: GON7, the fifth subunit of human KEOPS, bound to LAGE3-OSGEP (EKC/KEOPS complex subunit:Probable tRNA N6-adenosine threonylcarbamoyltransferase) Specimen concentration: 1.48 mg/ml / Entity id: 1759 / 1760 / 1761 |
|---|---|
| Buffer | Name: 20 mM MES, 200 mM NaCl, 5 mM β-mercaptoethanol / pH: 6.5 |
| Entity #1759 | Name: GON7 / Type: protein / Description: EKC/KEOPS complex subunit GON7 / Formula weight: 11.682 / Num. of mol.: 1 / Source: Homo sapiens / References: UniProt: Q9BXV9 Sequence: MELLGEYVGQ EGKPQKLRVS CEAPGDGDPF QGLLSGVAQM KDMVTELFDP LVQGEVQHRV AAAPDEDLDG DDEDDAEDEN NIDNRTNFDG PSAKRPKTPS HHHHHH |
| Entity #1760 | Name: LAGE3 / Type: protein / Description: EKC/KEOPS complex subunit LAGE3 / Formula weight: 14.804 / Num. of mol.: 1 / Source: Homo sapiens / References: UniProt: 1-143 Sequence: MRDADADAGG GADGGDGRGG HSCRGGVDTA AAPAGGAPPA HAPGPGRDAA SAARGSRMRP HIFTLSVPFP TPLEAEIAHG SLAPDAEPHQ RVVGKDLTVS GRILVVRWKA EDCRLLRISV INFLDQLSLV VRTMQRFGPP VSR |
| Entity #1761 | Name: OSGEP / Type: protein Description: Probable tRNA N6-adenosine threonylcarbamoyltransferase Formula weight: 36.426 / Num. of mol.: 1 / Source: Homo sapiens / References: UniProt: Q9NPF4 Sequence: MPAVLGFEGS ANKIGVGVVR DGKVLANPRR TYVTPPGTGF LPGDTARHHR AVILDLLQEA LTESGLTSQD IDCIAYTKGP GMGAPLVSVA VVARTVAQLW NKPLVGVNHC IGHIEMGRLI TGATSPTVLY VSGGNTQVIA YSEHRYRIFG ETIDIAVGNC LDRFARVLKI ...Sequence: MPAVLGFEGS ANKIGVGVVR DGKVLANPRR TYVTPPGTGF LPGDTARHHR AVILDLLQEA LTESGLTSQD IDCIAYTKGP GMGAPLVSVA VVARTVAQLW NKPLVGVNHC IGHIEMGRLI TGATSPTVLY VSGGNTQVIA YSEHRYRIFG ETIDIAVGNC LDRFARVLKI SNDPSPGYNI EQMAKRGKKL VELPYTVKGM DVSFSGILSF IEDVAHRMLA TGECTPEDLC FSLQETVFAM LVEITERAMA HCGSQEALIV GGVGCNVRLQ EMMATMCQER GARLFATDER FCIDNGAMIA QAGWEMFRAG HRTPLSDSGV TQRYRTDEVE VTWRD |
-Experimental information
| Beam | Instrument name: SOLEIL SWING  / City: Saint-Aubin / 国: France / City: Saint-Aubin / 国: France  / Type of source: X-ray synchrotron / Wavelength: 0.1 Å / Dist. spec. to detc.: 1.8 mm / Type of source: X-ray synchrotron / Wavelength: 0.1 Å / Dist. spec. to detc.: 1.8 mm | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Detector | Name: AVIEX PCCD170170 / Type: CCD | |||||||||||||||||||||||||||||||||
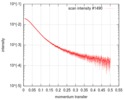
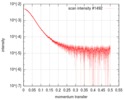
| Scan | Measurement date: May 12, 2017 / Cell temperature: 15 °C / Exposure time: 1 sec. / Number of frames: 255 / Unit: 1/A /
| |||||||||||||||||||||||||||||||||
| Distance distribution function P(R) |
| |||||||||||||||||||||||||||||||||
| Result | Comments: The scattered intensities were displayed on an absolute scale (cm-1) using the scattering of water. SAXS data were normalized to the intensity of the incident beam, averaged and background ...Comments: The scattered intensities were displayed on an absolute scale (cm-1) using the scattering of water. SAXS data were normalized to the intensity of the incident beam, averaged and background subtracted using the program US-SOMO. Frames were examined individually and 21 identical frames were averaged and further processed. The corresponding SEC-elution concentration was approximately 0.35 mg/mL. The complex is constituted of one monomer of OSGEP, one monomer of LAGE3 and one monomer of GON7. Molecular mass was obtained using the Bayesian inference approach proposed in PRIMUS/qt, which combines four concentration independent MM estimators: M=59.5 kDa within the credibility interval [57.5-64.5].
|
 Movie
Movie Controller
Controller

 SASDFL8
SASDFL8